Abstract
In late 2019, an outbreak of pneumonia caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) initiated in Wuhan, Hubei province, China. The major clinical symptoms described for coronavirus disease (COVID-19) include respiratory distress and pneumonia in severe cases, and some patients may experience gastrointestinal impairments. In accordance, viral RNA or live infectious virus have been detected in feces of patients with COVID-19. Binding of SARS-CoV-2 to the angiotensin‐converting enzyme 2 (ACE2) is a vital pathway for the virus entry into human cells, including those of the respiratory mucosa, esophageal epithelium as well as the absorptive enterocytes from ileum and colon. The interaction between SARS-CoV-2 and ACE2 receptor may decrease the receptor expression and disrupt the function of B0AT1 transporter influencing the diarrhea observed in COVID-19 patients. In this context, a fecal-oral transmission route has been considered and points out a role for the digestive tract in disease transmission and severity. Here, in order to further understand the impact of COVID-19 in human physiology, the cellular and molecular mechanisms of SARS-CoV-2 infection and disease severity are discussed in the context of gastrointestinal disturbances.
In December 2019, an outbreak of coronavirus disease (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a positive‐sense single‐stranded RNA virus, was reported as a public health emergency of international concern [1]. First described as a disease of the respiratory tract, COVID-19 has rapidly spread from Wuhan, China to all over the world affecting more than 22 million people across 215 countries [2].
SARS-CoV-2 uses the metallopeptidase angiotensin-converting enzyme 2 (ACE2) as a viral receptor to enter the human cells. Coronavirus fusion with the cell membrane and internalization also depends on the interaction with the cellular serine protease transmembrane protease serine 2 (TMPRSS2), the cysteine proteases cathepsin B and L, furin and furin-like proteins [3]. As previously observed for SARS-CoV in 2003, and Middle East respiratory syndrome-CoV (MERS-CoV) in 2012 [4], SARS-CoV-2 is thought to be transmitted by respiratory droplets or aerosols and direct contact [5]. Once transmitted to an individual, typical presentations of the infection, such as dry cough, headache, fever, fatigue and pneumonia, are observed [6]. However, disease transmission and, the viral target cells and organs are still under investigation. Interestingly, it has recently been debated that some patients present with gastrointestinal manifestations, including diarrhea, nausea and vomiting. Some studies have also suggested that the gastrointestinal symptoms represent the onset manifestations of the disease [7]. Furthermore, viral RNA has been detected in COVID-19 patients stool samples [8] and rectal swabs [9] even when the virus is no longer found in the upper respiratory tract. These intriguing findings suggest that COVID-19 gastrointestinal symptoms may result from the interaction of SARS-CoV-2 with cells of the gastrointestinal tract as ACE2 is not only expressed in human alveolar epithelial cells but is also highly expressed in the oral mucosa [10], esophagus and stomach [11], small intestine and colon enterocytes [12]. From this perspective, the fecal-oral transmission may represent an important route of virus transmission. Additionally, the long-term changes that SARS-CoV-2 infection can cause in the gastrointestinal tract in previously healthy patients and in those who already present gastrointestinal diseases must also be considered.
ACE2 was first described as the human homolog of zinc metallopeptidase ACE. The enzyme negatively regulates the rennin-angiotensin system, which controls multiple body functions by converting angiotensin II (Ang II) into its lung-protective metabolite Ang-(1-7). ACE2 is a type 1 transmembrane protein anchored on the cell surface with a long extracellular N-terminal domain, that exhibits carboxymonopeptidase activity; a transmembrane region, and a short C-terminal cytoplasmic tail. The extracellular domain contains a single zincbinding motif, that shares about 42% sequence homology with the amino-terminal domain of ACE and forms the active site of the enzyme [13]. ACE2 was also identified as the functional receptor for the human pathogenic coronaviruses SARSCoV, HCoV-NL63 (human coronavirus NL63), and the new SARS-CoV-2 [14]. It was recently demonstrated by cryogenic electron microscopy that SARS-CoV-2 S protein binds to ACE2 with 10- to 20-fold affinity when compared to SARS-CoV [15].
ACE2 is highly expressed in many tissues, including the lung, heart, blood vessels, kidney, and testis (Table 1). In the context of COVID-19, ACE2 expression in lung AT2 cells favors airway infection and explains the high incidence of pneumonia in those with a severe disease course. Additional evidence indicates that ACE2 and TMPRSS2 are co-expressed in the upper epithelial cells of the esophagus and absorptive enterocytes, and that this expression pattern may facilitate the gastrointestinal infection [16]. Similarly, dipeptidyl peptidase-4 (DPP4), the receptor that interacts with MERS-CoV S protein, displayed similar distribution to that of ACE2 in the small intestine. DPP4-expressing human intestinal cells were highly susceptible to MERS-CoV infection, and supported a robust viral replication, indicating that the human gastrointestinal tract may be an important site of infection [17].
Interestingly, gut ACE2 is an important regulator of amino acid transport. ACE2 operates as a “chaperone” for the membrane trafficking of B0 AT1, the main sodium-dependent neutral amino acid transporter found in the kidney proximal tubule and small intestine. It was suggested, using ACE2 knockout mice (ACE2 KO), that ACE2 is essential for B0AT1 expression in the luminal surface of intestinal epithelial cells and pivotal to its stabilization in the small intestine [16,18]. ACE2 KO are completely absent of B0AT1 in the small intestine but exhibit normal expression of the transporter in the kidney [18]. This particular evidence suggests a tissue-specific role for B0AT1 and indicates that the expression and function of B0AT1 in the small intestine depends on its co-expression with ACE2 (Table 1) [18].
Mutations in the gene slc6a19 which encodes B0AT1 have been linked to the Hartnup disorder, an autosomal recessive disease associated with dermatological, neurological and psychiatric abnormalities due to defective amino acid absorption in the kidney and small intestine [21]. Of note, during infection, Hartnup sufferers can present pellagra-like symptoms such as emotional instability, cerebellar ataxia, as well as diarrhea [22]. ACE2 KO present with reduced amino acid uptake associated with diarrhea and ulcerative colitis susceptibility [23]; these, are frequently described in Hartnup and Pellagra patients. ACE2- deficient mice also exhibit reduced plasma levels of tryptophan, altered microbiota, and increased susceptibility to ulcerative colitis and severe diarrhea; indicating that a defective intestinal tryptophan absorption favors diarrhea and intestinal inflammation [23].
Although the knowledge of the specific mechanisms involved in the gastrointestinal symptoms of COVID-19 is still evolving, ACE2 expression and its relevance in the gastrointestinal tract suggest that SARS-CoV-2 is likely to cause both acute and long-term gastrointestinal alterations.
Coronavirus infections have been frequently linked to gastrointestinal alterations. Diarrhea was detected in up to 10% of SARS-CoV-positive patients [24]. Pathological studies showed that SARS-CoV infects not only the respiratory epithelium but also the surface enterocytes in the small intestine, favoring intestinal permeability to lipopolysaccharide and luminal bacteria [25]. SARS-CoV was also detected in stool specimen from patients with confirmed SARS infection for at least 26 days after onset of symptoms [26].
Nearly a quarter of the hospitalized MERS-CoV patients of a large cohort study performed in Saudi Arabia (between September 1, 2012 and June 15, 2013), presented with gastrointestinal manifestations, including abdominal pain (17%), vomiting (21%), and diarrhea (most common symptom, 26% of severe cases) [27]. Furthermore, it was also described that the first MERS-CoV patient, from France, initially presented with fever and diarrhea and no respiratory symptoms [28] suggesting that the gastrointestinal tract may serve as an alternative route for MERS-CoV infection.
The ability of MERS-CoV to infect intestinal cells was demonstrated by Zhou et al. [17], who showed that intestinal organoids, small intestine explants and, human primary intestinal epithelial cells are highly susceptible to MERS-CoV infection and maintain viral replication. Human epithelial colorectal adenocarcinoma cells were also susceptible to MERS-CoV infection [17]. The same study also showed that the intragastric inoculation of MERS-CoV in human DPP4 transgenic mice causes lethality, an effect which was associated with epithelial deterioration and inflammation in the small intestine. The enteric infection and inflammation progressively evolved to respiratory failure. The virus was found in stool specimens and although it lost its infectivity in highly acidic fasted-state gastric fluid, it was resistant to intestinal fluid and to a fed-state condition. As other enveloped coronaviruses, MERS-CoV was also resistant to bile salts [17]. These data suggest that the intestine is an important initial site of MERS-CoV infection and highlight the oral-fecal route as an essential transmission pathway of the disease. From this perspective, it is possible to infer that SARS-CoV, MERS-CoV and probably SARS-CoV-2, are clearly not just lung pathogens. Data regarding SARS-CoV-2 infection and gastrointestinal alterations will now be discussed.
Epidemiological studies indicated the presence of SARS-CoV-2 RNA in feces of 41 (55%) of 74 COVID-19 patients even after hospital discharge, and 5 weeks following respiratory samples became negative [8], with no differences between severe and non-severe clinical cases [24]. In another report, 39 out of the 73 hospitalized patients infected with SARS-CoV-2, tested positive for virus RNA in their stools for a period of 12 days; and 17 of the patients continued to exhibit positive stool samples after becoming negative in the nasopharyngeal tests [29].
However, it is still unclear if the digestive symptoms are primary or secondary consequences of SARS-CoV-2 infection. In fact, diarrhea may be the first symptom before diagnosis, or may appear after disease confirmation, Fang et al. [30] found that up to 49% of the patients experience diarrhea after hospital admission and antiviral treatment, while up to 22% of patients reported diarrhea before diagnosis. In addition to diarrhea, the gastrointestinal symptoms included, nausea, vomiting, abdominal pain, anorexia, and gastrointestinal bleeding [31,32].
Although beta-coronaviruses are not able to replicate outside of the host cell, they can survive for a long time (MERS-CoV: 48 hours; SARS-CoV and SARS-CoV-2: 72 hours) in dried surfaces at 21°C to 23°C and 40% relative humidity, and for up to 2 hours in aerosols [5]. Data obtained from a study performed in a hospital in Beijing, China, during the SARS-CoV outbreak in 2003, revealed that the virus remains infectious for 14 days in sewages at 4°C and 2 days at 20°C; and that viral RNA can be detected even 8 days following SARS-CoV inactivation [33]. Surprisingly, an investigation in Hong Kong suggested an untypical viral spread between residents of an apartment complex through sewer gases during the SARS-CoV outbreak [34]. Similarly, SARS-CoV-2 was also found in wastewater samples of various countries [35-37]. In this context, some authors have suggested that wastewater-based epidemiology would be an useful tool for monitoring infectious diseases such as COVID-19 [38]. Considering all the discussed aspects and the similarity between the coronaviruses, the possibility of a fecal-oral transmission of SARS-CoV-2 should be considered and gastrointestinal alterations should be better explored.
As mentioned before, a high frequency of gastrointestinalrelated symptoms has been observed in COVID-19 patients [39]. Clinical data from 1,099 hospitalized patients and outpatients infected with SARS-CoV-2 of 552 hospitals located at 30 different provinces of China, demonstrated that up to 5% of the subjects present with nausea or vomiting, and up to 4% had diarrhea [40]. In another study, 39% of 140 community‐positive infected COVID‐19 patients in Wuhan, China, complained of gastrointestinal symptoms, including poor appetite, abdominal pain, nausea, and diarrhea [41]. A study conducted in the Zhejiang province showed that up to 11% of the patients presented with at least one digestive symptom, including nausea, vomiting and diarrhea [32].
Interestingly, intestinal epithelial cells are susceptible to SARS-CoV-2 infection. Recent data demonstrated that enterocytes are rapidly infected by SARS and SARS-CoV-2 due to the presence of ACE2 and TMPRSS2 on the apical surface of these cells [42], suggesting a novel physiological site for viral replication and the consequent development of gastrointestinal manifestations, spreading of the virus from the gut to the lung, and transmission of the virus [43].
Diarrhea stands out as the main gastric symptom among patients with COVID-19. Considering that SARS-CoV-2 depends on ACE2 internalization to infect host cells, a possible loss of B0AT1 function may occur, influencing the progression of the gastrointestinal symptoms observed in infected individuals (Fig. 1). An analysis of 31 studies and 3.335 patients, indicated that up to 13% of the COVID-19 patients (random metaanalysis effect) presented with diarrhea; of note, the prevalence of diarrhea in SARS-CoV and MERS-CoV individuals was of 12% and 23%, respectively [44]. Pan et al. [45], noted that the gastrointestinal symptoms are more pronounced as disease outcome worsens; indeed, those with gastrointestinal symptoms are more likely to spend longer periods at the hospital in comparison with those without such manifestations.
In severe COVID-19 cases, patients may exhibit esophagus ulcer, gastrointestinal bleeding, and multiple round herpetic erosions [24]. A gastrointestinal endoscopy performed on a patient showed the presence of infiltrating plasma cells and lymphocytes in the esophageal squamous epithelium, accompanied by interstitial edema in the lamina propria of the stomach, duodenum, and rectum, but no tissue damage [29]. On the other hand, an autopsy report showed degeneration, necrosis and shedding of the gastrointestinal mucosa of COVID-19 patient [46].
Accordingly, an epidemiological study with severe COVID-19 patients in Zhuhai, China, detected SARS-CoV-2 RNA in esophagus, stomach, duodenum and rectum specimens of 2 patients [24]. ACE2 and viral nucleocapsid protein were observed in the stomach, duodenum, and rectum gastrointestinal epithelial cells [47], and SARS-CoV-2 RNA was found in 28 fecal samples (66%) of 42 tested patients [8,9,29]. Surprisingly, the virus isolated from the stool samples of severe COVID-19 sufferers were viable and able to infect susceptible cells [48] suggesting that the digestive symptoms of SARS-CoV-2 infection may result from the contact of the virus with the gastrointestinal cells.
In contrast, Zang et al. [49] conducted a series of in vitro studies and suggested that SARS-CoV-2 can be rapidly inactivated by gastric and intestinal fluids. These findings have since been debated as in vivo differences in human gastrointestinal fluids between fasted or fed-state, or even the variability of intestinal pH should must been considered [50]. Furthermore, the authors did not consider the virus contact with the mucosal tissue of oral cavity, since it expresses high amounts of ACE2.
Although most of the studies on SARS-CoV-2 have focused in the adult population, some clinical reports indicate that children may present a distinct pattern of response. A study with COVID-19-positive children from Wuhan, showed that 105 out of the 244 investigated infected children presented with SARS-CoV-2 in their stools. These results are quite interesting as only 34 out of the 244 children exhibited gastrointestinal symptoms on hospital admission [51]. The authors suggested that SARS-CoV-2 detection in stool samples may not be due to gut infection, but it is rather related to the swallowing of lung mucus and catarrh. They also reinforce that a fecal-oral transmission remains a possibility, as the live virus is detected in stool samples and toilet areas.
In this context, the identification of asymptomatic COVID-19 sufferers or those with mild enteric symptoms would be essential to better understand the disease and to determine more appropriated intervention protocols to reduce healthcare-associated infection and spread of the virus.
The physiological changes caused by SARS-CoV-2 infection are yet to be clarified and the long-term consequences will be seen over time. Gastrointestinal alterations at any level may affect other functions of the organism. A possible fecal-oral transmission deserves detailed investigations and subsequent early clinical interventions should be implemented to avoid SARS-CoV-2 infection and improve disease prognosis.
Notes
Funding Source
This work was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq; 305676/2019-9 and 408053/2018-6) and Instituto de Pesquisa Pelé Pequeno Príncipe. Barbosa da Luz B, de Oliveira NMT, França dos Santos IW are PhD and master students and receive grant from the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES).
REFERENCES
1. Malik YS, Sircar S, Bhat S, et al. Emerging coronavirus disease (COVID-19), a pandemic public health emergency with animal linkages: current status update. Preprints [Preprint]. 2020 [cited 2020 Sep 4]. https://doi.org/10.20944/preprints202003.0343.v1.

2. Worldometers.info. COVID-19 coronavirus pandemic, 10 June [Internet]. c2020. [cited 2020 Jun 10]. https://www.worldometers.info/coronavirus/?utm_campaign=homeAdvegas1.
3. Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020; 181:271–280.

4. Yeo C, Kaushal S, Yeo D. Enteric involvement of coronaviruses: is faecal-oral transmission of SARS-CoV-2 possible? Lancet Gastroenterol Hepatol. 2020; 5:335–337.

5. van Doremalen N, Bushmaker T, Morris DH, et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N Engl J Med. 2020; 382:1564–1567.

6. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020; 395:497–506.

7. Song Y, Liu P, Shi XL, et al. SARS- CoV-2 induced diarrhoea as onset symptom in patient with COVID-19. Gut. 2020; 69:1143–1144.

8. Wu Y, Guo C, Tang L, et al. Prolonged presence of SARSCoV-2 viral RNA in faecal samples. Lancet Gastroenterol Hepatol. 2020; 5:434–435.

9. Chen Y, Chen L, Deng Q, et al. The presence of SARS-CoV-2 RNA in the feces of COVID-19 patients. J Med Virol. 2020; 92:833–840.

10. Hamming I, Timens W, Bulthuis ML, Lely AT, Navis G, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004; 203:631–637.

11. Zou X, Chen K, Zou J, Han P, Hao J, Han Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front Med. 2020; 14:185–192.

12. Guo YR, Cao QD, Hong ZS, et al. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak - an update on the status. Mil Med Res. 2020; 7:11.

13. Clarke NE, Turner AJ. Angiotensin-converting enzyme 2: the first decade. Int J Hypertens. 2012; 2012:307315.

14. Li W, Moore MJ, Vasilieva N, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003; 426:450–454.

15. Wrapp D, Wang N, Corbett KS, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020; 367:1260–1263.

16. Perlot T, Penninger JM. ACE2 - from the renin-angiotensin system to gut microbiota and malnutrition. Microbes Infect. 2013; 15:866–873.

17. Zhou J, Li C, Zhao G, et al. Human intestinal tract serves as an alternative infection route for Middle East respiratory syndrome coronavirus. Sci Adv. 2017; 3:eaao–4966.

18. Camargo SM, Singer D, Makrides V, et al. Tissue-specific amino acid transporter partners ACE2 and collectrin differentially interact with hartnup mutations. Gastroenterology. 2009; 136:872–882.

19. Xu H, Zhong L, Deng J, et al. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int J Oral Sci. 2020; 12:8.

20. Jando J, Camargo SMR, Herzog B, Verrey F. Expression and regulation of the neutral amino acid transporter B0AT1 in rat small intestine. PLoS One. 2017; 12:e0184845.

21. Seow HF, Bröer S, Bröer A, et al. Hartnup disorder is caused by mutations in the gene encoding the neutral amino acid transporter SLC6A19. Nat Genet. 2004; 36:1003–1007.

22. Chawla J, Kvarnberg D. Hydrosoluble vitamins. In: Biller J, Ferro JM, eds. Neurologic aspects of systemic disease part II. 1st ed. Amsterdam: Elsevier, 2014:891-914.
23. Hashimoto T, Perlot T, Rehman A, et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature. 2012; 487:477–481.

24. Lin L, Jiang X, Zhang Z, et al. Gastrointestinal symptoms of 95 cases with SARS-CoV-2 infection. Gut. 2020; 69:997–1001.

25. To KF, Tong JH, Chan PK, et al. Tissue and cellular tropism of the coronavirus associated with severe acute respiratory syndrome: an in-situ hybridization study of fatal cases. J Pathol. 2004; 202:157–163.

26. Isakbaeva ET, Khetsuriani N, Beard RS, et al. SARS-associated coronavirus transmission, United States. Emerg Infect Dis. 2004; 10:225–231.

27. Assiri A, Al-Tawfiq JA, Al-Rabeeah AA, et al. Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: a descriptive study. Lancet Infect Dis. 2013; 13:752–761.

28. Mailles A, Blanckaert K, Chaud P, et al. First cases of Middle East respiratory syndrome coronavirus (MERS-CoV) infections in France, investigations and implications for the prevention of human-to-human transmission, France, May 2013. Euro Surveill. 2013; 18:20502.

29. Xiao F, Tang M, Zheng X, Liu Y, Li X, Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020; 158:1831–1833.

30. Fang D, Ma J, Guan J, et al. A single-center, descriptive study of the digestive system performance of hospitalized patients with novel coronavirus pneumonia in Wuhan area. Chin J Digestion. 2020; 40:E005–E005.
31. Tian Y, Rong L, Nian W, He Y. Review article: gastrointestinal features in COVID-19 and the possibility of faecal transmission. Aliment Pharmacol Ther. 2020; 51:843–851.

32. Jin X, Lian JS, Hu JH, et al. Epidemiological, clinical and virological characteristics of 74 cases of coronavirus-infected disease 2019 (COVID-19) with gastrointestinal symptoms. Gut. 2020; 69:1002–1009.
33. Wang XW, Li J, Guo T, et al. Concentration and detection of SARS coronavirus in sewage from Xiao Tang Shan Hospital and the 309th Hospital of the Chinese People’s Liberation Army. Water Sci Technol. 2005; 52:213–221.

34. McKinney KR, Gong YY, Lewis TG. Environmental transmission of SARS at Amoy Gardens. J Environ Health. 2006; 68:26–30.
35. Randazzo W, Truchado P, Cuevas-Ferrando E, Simón P, Allende A, Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020; 181:115942.

36. La Rosa G, Iaconelli M, Mancini P, et al. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci Total Environ. 2020; 736:139652.

37. Ahmed W, Angel N, Edson J, et al. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community. Sci Total Environ. 2020; 728:138764.

38. Rimoldi SG, Stefani F, Gigantiello A, et al. Presence and infectivity of SARS-CoV-2 virus in wastewaters and rivers. Sci Total Environ. 2020; 744:140911.

39. Wong SH, Lui RN, Sung JJ. Covid-19 and the digestive system. J Gastroenterol Hepatol. 2020; 35:744–748.

40. Guan WJ, Ni ZY, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020; 382:1708–1720.

41. Zhang JJ, Dong X, Cao YY, et al. Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China. Allergy. 2020; 75:1730–1741.

42. Lamers MM, Beumer J, van der Vaart J, et al. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020; 369:50–54.

43. Stanifer ML, Kee C, Cortese M, et al. Critical role of type III interferon in controlling SARS-CoV-2 infection, replication and spread in primary human intestinal epithelial cells. BioRxiv [Preprint]. 2020 [cited 2020 Sep 4]. https://doi.org/10.1101/2020.04.24.059667.

44. Cheung KS, Hung IFN, Chan PPY, et al. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from a Hong Kong cohort: systematic review and metaanalysis. Gastroenterology. 2020; 159:81–95.

45. Pan L, Mu M, Yang P, et al. Clinical characteristics of COVID-19 patients with digestive symptoms in Hubei, China: a descriptive, cross-sectional, multicenter study. Am J Gastroenterol. 2020; 115:766–773.

46. Yao XH, Li TY, He ZC, et al. A pathological report of three COVID-19 cases by minimal invasive autopsies. Zhonghua Bing Li Xue Za Zhi. 2020; 49:411–417.
47. Uno Y. Why does SARS-CoV-2 invade the gastrointestinal epithelium? Gastroenterology. 2020; 159:1622–1623.

48. Xiao F, Sun J, Xu Y, et al. Infectious SARS-CoV-2 in feces of patient with severe COVID-19. Emerg Infect Dis. 2020; 26:1920–1922.

49. Zang R, Gomez Castro MF, McCune BT, et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci Immunol. 2020; 5:eabc3582.

Fig. 1.
Mechanism suggested for diarrhea induced by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. During coronavirus disease (COVID-19), SARS-CoV-2 uses the angiotensin-converting enzyme 2 (ACE2) receptor and cellular serine protease transmembrane protease serine 2 (TMPRSS2) for host cell entry. ACE2 is an important regulator of amino acid transport B0AT1. The interaction between SARS-CoV-2 and ACE2 receptor may decrease the receptor expression leading to B0AT1 disruption, influencing the diarrhea observed in COVID-19 patients.
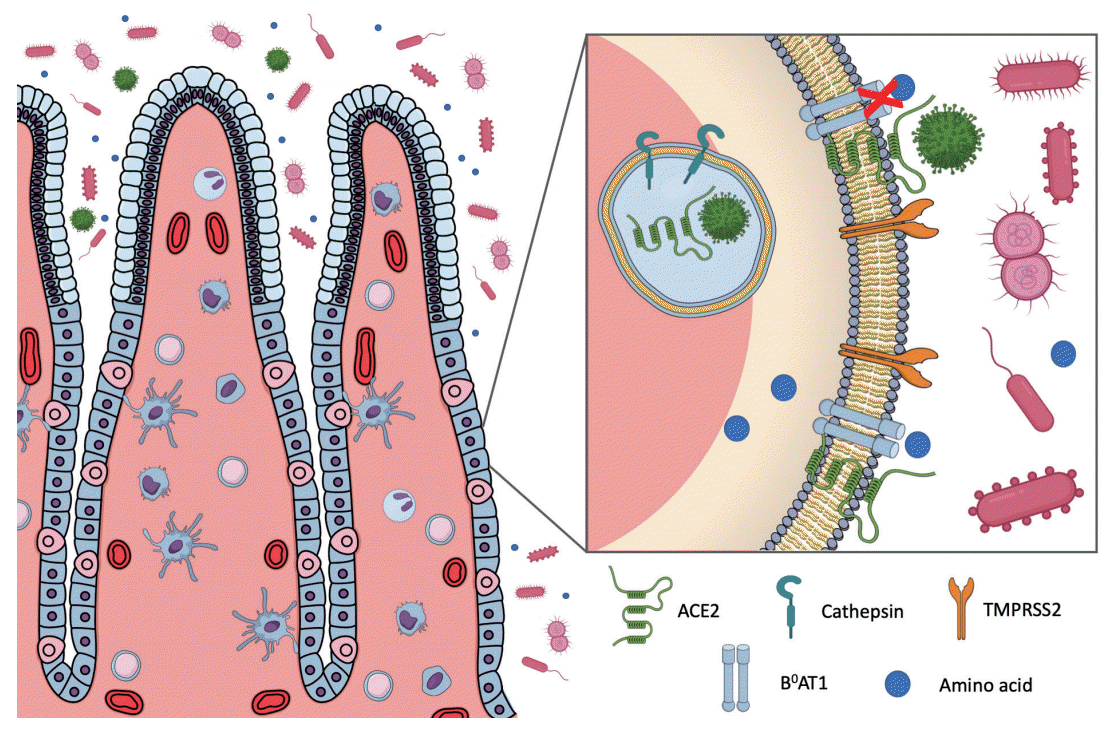
Table 1.
ACE2 and B0AT1 Tissue Expression
| Protein | Organ or tissue expression | Cell type | Assay | Involvement with COVID-19 | Author (year) |
|---|---|---|---|---|---|
| ACE2 | Esophagus | Epithelial cells | scRNA-seq | Contributes to virus spread and immunopathology in airways and intestinal system | Zou et al. (2020) [11] |
| Intestine | Enterocytes and intestinal villi | scRNA-seq | Indicates an intrinsic susceptibility of intestine to SARS-CoV-2 infection and replication | Zou et al. (2020) [11], Hamming et al. (2004) [10] | |
| Oral cavity | Epithelial cells | RNA-seq | Potential route of SARS-CoV-2 infection | Xu et al. (2020) [19], Zou et al. (2020) [11] | |
| Stomach | Epithelial cells | scRNA-seq | Potential route of SARS-CoV-2 infection | Zou et al. (2020) [11] | |
| Intestine | Enterocytes and intestinal villi | Western blot | Modulates the diarrhea that occurs in up to 39% of COVID-19 cases | Jando et al. (2017) [20] | |
| B0AT1 | Intestine | Enterocytes and intestinal villi | Western blot | Modulates the diarrhea that occurs in up to 39% of COVID-19 cases | Jando et al. (2017) [20] |




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download