Abstract
Inflammatory bowel diseases (IBD) are chronic conditions of the gastrointestinal tract-the main site of host-microbial interaction in the body. Development of IBD is not due to a single event but rather is a multifactorial process where a patient’s genetic background, behavioral habits, and environmental exposures contribute to disease pathogenesis. IBD patients exhibit alterations to gut bacterial populations “dysbiosis” due to the inflammatory microenvironment, however whether this alteration of the gut microbiota precedes inflammation has not been confirmed. Emerging evidence has highlighted the important role of gut microbes in developing measured immune responses and modulating other host responses such as metabolism. Much of the work on the gut microbiota has been correlative and there is an increasing need to understand the intimate relationship between host and microbe. In this review, we highlight how commensal and pathogenic bacteria interact with host intestinal epithelial cells and explore how altered microenvironments impact these connections.
Inflammatory bowel diseases (IBD), such as CD and UC, are complex, multifactorial disorders characterized by chronic inflammation of the GI tract. In the United States alone, it is estimated that IBD affects up to 1.3% of the population (3 million individuals) and a further 2.2 million individuals in Europe, with incidence rising in newly industrialized countries (www.cdc.gov/ibd/data-statistics.htm) [1-3]. Our current understanding suggests IBD manifests largely in genetically susceptible individuals following some perturbation of mucosal homeostasis which permits commensal gut flora to activate inflammatory processes beyond homeostatic levels. In agreement with this hypothesis, studies in gnotobiotic mice have demonstrated that transgenic mouse models of colitis require a microbial community to develop colitis [4,5]. In addition, administration of antibiotic therapy in IBD patients has been shown to induce remission further demonstrating that gut microbes promote disease [6]. Together, these studies point to the importance of gut flora in the pathogenesis of IBD.
The gut microbiota consists of bacteria, viruses, archaea and eukaryotic microbes. Bacteria constitute by far the largest component and both beneficial bacteria “commensals” and opportunistic pathogenic bacteria “pathobionts” are found within the GI tract. Gut bacterial numbers roughly equal the number of intestinal epithelial cells (IECs) and represent a large reservoir of gene expression that can affect host processes essential to GI function [7]. Indeed, the co-evolution of host and microbiota supports the notion that gene expression of the latter can affect the host [8]. The most prominent populations of gut flora are present in the large intestine, where bacteria play a critical role in the digestion and absorption of urea, bile acids, sterols, xenobiotics, essential vitamins, plant-based polysaccharides, and certain amino acids. The resulting metabolites are capable of altering IEC signaling and gene expression. In addition to their digestive function, bacteria within the gut are also important for maturation of the immune system and can modulate the immune response by both promoting certain functions and triggering anti-inflammatory responses [9-12].
The composition of the microbiota is fluid as alterations in diet and the presence of non-infectious disease have been shown to shift certain subpopulations within the gut microbiota [13,14]. These resulting communities can also induce phenotypic traits when administered to gnotobiotic mice [13]. Additionally, various changes in the features of the intestinal epithelium during IBD pathogenesis promote the proliferation and dominance of pathobionts such as adherent-invasive Escherichia coli (AIEC). However, whether changes in microbial populations precede disease development onset is still under investigation. Several studies have attempted to understand the role(s) of the microflora in disease pathogenesis by investigating specific bacterial shifts associated with IBD (specifically the prevalence of Enterobacteriaceae and Proteobacteria in IBD patient intestinal tissues) and the effect of IBD candidate genes on the interaction between IECs and the microflora.
In this review, we will summarize the present understanding of how the interaction of IECs and gut microbiota can either maintain healthy homeostasis or lead to dysbiosis by highlighting specific examples of commensals and pathobionts. We focused attention on the interactions of IECs with luminal bacteria as the interplay of the gut microbiota and mucosal immune cells has been comprehensively described previously [10,15,16].
In order to understand the impact of the gut microbiota in intestinal diseases we must first consider the major site of interaction: the intestinal barrier. The broader interpretation of the intestinal barrier requires the interplay of multiple components: IECs, the gut microbiota, and mucosal and submucosal immune cells. More specific barrier functions such as permeability to electrolytes and luminal contents are controlled directly by epithelial cells and their key structural components that regulate paracellular permeability, the apical tight junctions. For clarity, we will discriminate between permeability.referring to specific tight junction modifications.and the broader concept of barrier function in the following sections.
The intestinal epithelium is a continuous monolayer of specialized cells generated by intestinal stem cells at the base of the crypt. IEC differentiation from the crypt to the villus tip (small intestine), or surface epithelium (large intestine), and their subsequent turnover is critical for the chemical and physical functions of the barrier. IEC apoptosis and shedding into the lumen is tightly regulated to promote continuous turnover of the epithelium and maintain barrier integrity [17-19]. In a healthy gut, only 1% to 2% of the epithelia are undergoing apoptosis at any one time, and neighboring IECs act to seal gaps arising from apoptosis or cell “shedding events” in order to prevent unregulated access across the epithelium by luminal contents or microbes. Indeed, the pro-inflammatory cytokine TNF-α increases the number of shedding events however, this is not associated with a barrier defect due to the “gap-sealing” actions of adjacent epithelial cells [18,19]. The life cycle of an IEC is driven by changes in enterocyte function and epithelial differentiation. Villus or surface enterocytes display an absorptive phenotype while IECs at the crypt base secrete Cl-ions and water that aid in hydrating the mucus layer and flushing bacteria out of the crypt [20]. These functions are critical for the principal roles of the intestine (digestion, nutrient absorption, and waste excretion) and maintaining balanced interactions with luminal bacteria.
In addition to their absorptive and secretory functions, IECs also differentiate into specialized cells that contribute to intestinal barrier homeostasis. Paneth cells that are situated in the crypt base of the small intestine secrete anti-microbial factors such as α-defensins (or cryptidins in mice), RegIIIγ, lysozyme, and phospholipase A2 in response to microbial components [21]. These secreted factors aid in preventing pathogenic bacterial attachment and promote growth of beneficial bacteria. Positioned along the length of crypts, goblet cells secrete mucin proteins to establish a protective mucus layer overlying the intestinal epithelium that also functions as a diffusive medium for metabolites and nutrients. The composition and physical properties of the mucus layer differ by location in the GI tract. The small intestine has a loose, non-adherent mucus layer which facilitates nutrient absorption. In the colon, the mucus layer is composed of 2 sublayers–an outer layer much like that of the small intestine and a thick inner adherent layer that is impermeable to bacteria in the distal colon and somewhat permeable in the proximal colon [22]. Commensal microbes aid in maintaining this mucus layer through stimulation of mucin secretion. This observation has also been supported by analysis of the mucus layer in germ-free mice where the colonic inner layer was less developed [23]. Finally, the mucus binding capacity of probiotic bacteria is correlated with their colonization capacity, providing a selective advantage over pathogenic bacteria [24]. Studies on cystic fibrosis transmembrane conductance regulator knockout mice, which display accumulations of mucus, have shown an increase in IEC inflammatory gene expression and bacterial overgrowth, suggesting an abnormal mucus layer can promote dysbiosis [25,26].
A primary function of IECs is their ability to act as mediators of innate immunity. Similar to other cell types, IECs express cell-surface and intracellular pattern recognition receptors such as Toll-like receptors (TLRs) and NOD-like receptors, respectively. TLRs expressed by IECs function to respond primarily to commensal bacteria and different regions of the intestinal tract exhibit distinct patterns of TLR expression [27]. Accordingly, TLR expression is highest in the colon where their activation promotes expression of host defense genes [27]. In addition, the presence of gut flora or inflammation can also alter the expression profile of IEC TLRs. Activation of TLRs by commensal bacteria promotes tight junction protein expression to fortify the barrier and increases secretion of anti-microbial peptides [28-30].
Immune cells located in the mucosa and submucosa also function in shaping the microbial environment and hampering colonization by pathogenic bacteria. Lymphoid follicles called Peyer’s patches are located at various intervals in the ileum and consist of macrophages, lymphocytes, and M or micro-fold cells. These sites function in maintaining immune tolerance of commensal bacteria. In addition, secretion of IgA and components of the complement system aid in inhibiting pathogenic bacterial growth. Finally, certain bacteria are also necessary for immune maturation, such as segmented filamentous bacteria which promote Th17 cell development and Bacteroides fragilis which influences regulatory T cell development, and Th2 and Th1 cell balance [31-38].
Secretory IgA is a mucosal Ig produced by intestinal lamina propria B cells that captures microbial pathogens and their toxins to prevent adherence and invasion. Induction of IgA secretion occurs at Peyer’s patches and isolated lymphoid follicles within the GI wall where signals from T cells, dendritic cells, and IECs cause class switching of B cells to IgA-producing plasma cells [39]. Importantly, IgA is able to protect the barrier from bacteria without increasing inflammation. Commensal bacteria can promote IgA production directly through bacterial proteins binding host receptors on IECs, and indirectly through microbial metabolites such as short-chain fatty acids (SCFAs) binding the GPR43 receptor [40,41]. In turn, IgA binding to commensals can promote their colonization and stability within their colonization niche [42-44]. A continuing question in the field is how host responses differentiate between commensal and pathogenic microorganisms. It is thought that IgA binding to commensals is low-affinity whereas increased coating of bacteria by IgA indicates the increased pathogenic potential of the bacterium. This has been supported by transfer of highly IgA-coated bacteria isolated from IBD patients to germ-free mice which led to increased susceptibility to dextran sulfate sodium (DSS) colitis [45]. Thus, IgA is an important discriminator of commensal and pathogenic bacteria and promotes mucosal homeostasis through increased colonization of healthy commensals.
Normal antigen-sampling of the GI luminal contents by IECs and submucosal immune cells elicits a tolerogenic immune response essential for shaping GI microbial composition. The role of IECs as mediators of innate immunity is underscored by studies of single-nucleotide polymorphisms (SNPs) associated with IBD, in which genes critical for antigen recognition and bacterial destruction have been identified, such as nucleotide-binding oligomerization domain 2 (NOD2) and autophagy-related protein 16-like 1 (ATG16L1) [46-50]. Deficiency of these genes impairs normal maintenance of commensal populations and restriction of pathobionts, leading to dysbiosis [51-55]. The influence of IBD candidate genes on epithelial barrier function and bacterial sensing are discussed in greater detail in several comprehensive review articles [56,57].
Commensal microbes are those normally present in a healthy gut and who contribute to host mucosal homeostasis. In addition to their critical roles in digestion for the host, they also serve to support the intestinal barrier by promoting tolerogenic immune responses, as alluded to earlier. In particular there are a number of commensals that have been well-studied for their ability to improve barrier function and promote intestinal health. In this section, we review a number of these commensal organisms in an effort to understand how microbes contribute to maintenance of the intestinal epithelial barrier.
The mucus layer is composed of mucins with oligosaccharide chains whose terminal ends cannot be metabolized by the majority of gut bacteria [58]. However, certain bacteria termed mucin specialists have evolved specific enzymes that allow degradation of mucin oligosaccharide chains for sugar and protein, thereby facilitating their own colonization as well as growth of other commensals by increasing nutrient availability [59-62]. In fact, commensals such as Faecalibacterium prausnitzii rely on these mucin specialists to facilitate their colonization to the mucus layer [63-68]. Akkermansia muciniphila is a widely studied mucin specialist capable of tolerating the hypoxic environment near the epithelial surface and that utilizes host-derived proteins for survival [69-71]. A. muciniphila colonization increases mucin production and mucus thickness, and improves barrier function [70,72-74]. Butyrate production by A. muciniphila is also beneficial to the host by upregulating energetic pathways, such as increased β-oxidation and intestinal gluconeogenesis in colonocytes that maintain a commensal-dominated microenvironment and protect from metabolic impairment [75-78]. Finally, increased abundance of A. muciniphila at sites of intestinal injury where oxygen is depleted, allows expanded colonization by anaerobic bacteria. In this setting, A. muciniphila also promotes intestinal epithelial formyl peptide receptor 1 (FPR1) signaling and NADPH oxidase (NOX1) activity to increase enterocyte migration and proliferation to facilitate mucosal healing [79].
In addition, A. muciniphila has been inversely associated with intestinal inflammation [62,80-84]. Recent evidence has also shown that A. muciniphila-secreted extracellular vesicles protect the epithelium from colitis and diet-induced barrier dysfunction [85]. Another muciniphile Peptostreptococcus russellii, is also protective against DSS-induced chemical injury colitis model in mice. In addition to its mucin-degrading activity, P. russellii expresses aromatic amino acid metabolic enzymes that utilize tryptophan to produce indoleacrylic acids, which have anti-inflammatory effects and promote goblet cell function [86]. The phenyllactate dehydratase gene cluster responsible for expression of these enzymes was decreased in IBD patients compared to healthy controls [86]. Collectively, these findings indicate both host and commensal act synergistically to encourage mucosal homeostasis and repress inflammation.
Another commensal microbe that is consistently decreased in UC patients is F. prausnitzii [87-89]. F. prausnitzii is a highly abundant butyrate-producer in the intestinal tract, encompassing 5% of bacteria in feces [90,91]. Since increased F. prausnitzii is correlated with improved outcomes following surgical resection, it was thought that this microbe could directly affect host immunity although its presence is not required for patient recovery [92,93]. In accordance with this, Quevrain et al. [94] identified a 15 kDa anti-inflammatory protein was secreted by F. prausnitzii. Subsequently, a study by Breyner et al. [95] demonstrated F. prausnitzii secreted peptides termed “microbial anti-inflammatory molecules” can inhibit nuclear factor (NF)-κB promoter activity and interleukin (IL)-6 production, leading to protection from experimental colitis in mice through alteration of T cell immune responses. Interestingly, co-culture studies have demonstrated F. prausnitzii does not enhance barrier function but can induce activation of NF-κB signaling downstream of IL-1β in Caco-2 cells [92,96]. In contrast, Caco-2 cells treated with F. prausnitzii supernatant decreases basal and IL-1β-induced NF-κB activity [92].
L. rhamnosus Gorbach-Goldin (LGG) has been well-studied as a probiotic used in the production of dairy yogurt. This commensal microbe can indirectly and directly regulate epithelial function to repress inflammation and resist enteric infections. Administration of LGG culture supernatant to neonatal rats was associated with increased IEC proliferation, decreased paracellular barrier permeability, and increased expression of mucin-2, zonula occludens-1 (ZO-1), and IgA, together promoting resistance to neonatal E. coli K1 infection [97]. In addition, LGG enhances wound healing through enterocyte activation of FPR1 and NOX1, similar to A. muciniphila [98]. Direct actions of LGG are performed p40, were found to inhibit pro-inflammatory cytokine-induced IEC apoptosis [99]. In addition, p40 can also modulate IgA production by increased expression of a proliferation-inducing ligand in IECs [41]. More recently, p40 was found to exert its responses through the epidermal growth factor receptor in neonatal mice, ultimately promoting IgA production and T regulatory cell (Treg) differentiation in adult mice [100].
B. fragilis is a commensal found within the glycocalyx and in colonic crypts of mice that exhibits single-strain stability or the ability of a single strain of bacteria to persist in the microbial community for years [44]. Its colonization of the mucosa is facilitated by IgA binding, which allows the bacteria to aggregate and contribute to colonization resistance against pathogenic bacteria [44]. In addition, its expression of cytochrome bd oxidase permits its growth in low concentrations of oxygen, such as those present in the colon [101]. B. fragilis secretes an immunomodulatory molecule, polysaccharide A (PSA) that regulates immune cells such as induction of Foxp3+ Treg cell differentiation to reduce intestinal inflammation and repression of IL-17 secretion [36-38]. Furthermore, B. fragilis strains expressing PSA are able to prevent colonization by the pathobiont Helicobacter hepaticus and subsequent inflammation, indicating commensals can prevent pathobiont expansion [37]. Intriguingly, B. fragilis also accounts for a majority of Bacteroides infections in the body and the enterotoxigenic B. fragilis (ETBF) strain is a causative pathogen of diarrhea [102]. Recently, Chan et al. [103] demonstrated stable colonization of non-enterotoxigenic B. fragilis (NTBF) ameliorated disease severity following ETBF infection, in a PSA-independent manner. This study confirmed that B. fragilis predominance alone can be protective against mucosal inflammation. Whether increased virulence results from changes in the GI microenvironment and microbial exchange of genetic information is not well understood, although this is an emerging topic among research groups [102].
Inflammation in the GI tract is able to both alter the microbial community composition and promote expansion of pathogenic bacteria, as described in extensive detail elsewhere [14-16]. Metagenomic studies of fecal matter, intestinal wash, and intestinal tissues have observed reduced diversity and temporal instability of the microflora in IBD patients compared to healthy controls despite an increased number of microflora at involved tissue sites in IBD patients [104]. Bacterial communities can be influenced by inflammation-induced alterations of nutrient sources, luminal pH, and growth of other bacterial species such as those producing SCFAs which can also affect luminal pH due to their acidic profile. In some cases, inflammation can support the virulent activity of commensals known as pathobionts [105-107]. For example, Chassaing and Gewirtz [108] demonstrated that expansion of AIEC in mice lacking TLR5 required a pro-inflammatory niche. Here we summarize how inflammation-induced changes in mucosal homeostasis, particularly the mucosal microenvironment, promotes dysbiosis (Table 1) [78,79,109-118].
Under healthy conditions, an oxygen gradient in the colonic mucosa exists as a result of the high oxygen consumption required to maintain the inward Na+ gradient driving fluid absorption. However, bacteria can also contribute to the colonic oxygen gradient. Butyrate-producing bacteria increase levels of CO2 and together these mechanisms promote a hypoxic environment in the colonic lumen. Decreased levels of O2 promote anaerobic growth of commensal bacteria such as B. fragilis and F. prausnitzii closer to the surface of IECs [101,110]. However, in a chronic inflammatory state, oxygen levels near IECs increase due to secretion of reactive oxygen species and increased blood flow. This allows facultative anaerobic, or “aerotolerant,” bacteria to predominate near the intestinal epithelium such as Actinobacteria and Proteobacteria, phyla that are also associated with IBD [119]. For example, mice exposed to DSS had increased numbers of aerobic bacteria compared to healthy controls, a phenomenon that has also been observed in newly-diagnosed CD patients [109,120]. Depletion of butyrate-producers such as Clostridia by antibiotic therapy induces a subsequent increase in luminal oxygen, allowing increased growth of Enterobacteriaceae such as Salmonella serovar typhimurium [121]. The concept of increased epithelial oxygenation acting as a driver of facultative anaerobe expansion at the expense of butyrate-producing obligate anaerobes is elegantly described in a recent review by Litvak et al. [78]
A primary method of preventing pathobiont expansion is colonization resistance by homeostatic microbes. Efficient nutrient assimilation provides a selective advantage to certain bacteria that can hinder growth of pathogens. In addition, metabolites in the GI lumen can also activate signaling processes within IECs that promote selective microbial growth in both the healthy and inflamed gut. During inflammation, secreted reactive nitrogen species such as nitric oxide degrade to nitrate in the lumen where they are utilized by microbial nitrogen respiration pathways. Similarly, production of reactive oxygen species promotes alternative sources of electron acceptors that are then utilized in microbial respiration. For example, hydrogen sulfide (H2S) produced by commensals is oxidized to thionate during inflammation which is then utilized by S. typhimurium, although there has been some evidence indicating H2S is also anti-inflammatory and promotes mucosal healing [111,122]. Another metabolite increased by intestinal inflammation includes ethanolamine (EA), which is produced from phosphatidylethanolamine. Enzymes that utilize EA as a nutrient source are found in the genomes of many enteric pathogens so it is unsurprising that EA promotes predominance of S. typhimurium and enterohemorrhagic E. coli in mice [112,123]. Similarly, the bacterial fermentation product 1,2-propanediol is exploited by S. typhimurium during inflammation with increased expression of virulence factors [124]. Finally, GPR43, a Gprotein coupled receptor bound by bacterial-produced SCFAs, is important for resolution of intestinal inflammation in mice, and thus indicates a link between gut microbiota and inflammation resolution [125].
Availability of dietary transition metals can also restrict pathogenic bacteria growth. In a recent study Zhu et al. [114] confirmed that microbial molybdenum cofactor-dependent respiration is a metabolic signature of DSS-induced dysbiosis and promotes Enterobacteriaceae expansion during colitis [126]. Consistently, pathobiont expansion was reversed by tungstate treatment [114]. Competition for iron by the E. coli strain Nissle 1917 restricts S. typhimurium during colitis and infection [115]. Interestingly, the host protein lipocalin-2 is required for this effect, indicating an intricate interaction between host and commensal bacteria that promotes a healthy gut environment. In addition to colonization resistance, dietary metals can also alter bacterial gene expression. Zinc deficiency elevates the risk of pediatric diarrhea and is associated with pathogenic E. coli. [127]. Indeed, low zinc causes increased virulence of enteroaggregative E. coli, the pathogen responsible for traveler’s diarrhea [116].
Another factor affecting the gut microbial community is luminal pH, which can range from 5.7 to 6.8 in human cecum and pH 6.1 to 7.5 in the colon [128]. There are major shifts in the gut microbiota between luminal pH values of 5.5 and 6.5. In a healthy colon, a gradient from a mildly acidic to a neutral environment exists from the proximal to distal colon. The acidic pH in the proximal colon is maintained by substrate fermentation where acetate and other SCFAs are produced from the fermentation of indigestible polysaccharides facilitating expansion of butyrate-producing species such as Roseburia [129,130]. During spontaneous colitis, accumulation of lactate drives a decrease in luminal pH, potentially due to repressed growth of lactateutilizing bacteria [131,132].
From these examples, it is clear that environmental changes of the microbial niche can alter composition of the gut microbiota, occurring as both small perturbations and large displacement of commensal bacteria. Both diet and inflammatory status contribute to these effects. Apart from these shifts in population, changes in pathogenicity of seemingly innocuous bacteria are also of great interest since chronic GI conditions are associated with increased numbers of more virulent bacteria including “pathobionts.”
Pathobionts are bacteria normally found in the intestines that exhibit pathogenic activity during a chronic disease state or in a genetically-susceptible host [133]. Examples of pathobionts include Helicobacter species, virulent strains of E. coli, Campylobacter concisus, Klebsiella species, and Vancomycin-resistant Enterococcus faecalis, among others. These bacteria promote inflammation by disruption of the intestinal barrier, invasion of the epithelium, and/or modulation of inflammatory responses. Notably, in mouse models monoassociated with pathobiont bacteria increased disease severity is dependent upon the presence of resident microbiota [134]. In addition, activated immune cell responses against both resident and pathogenic bacteria suggest that background inflammatory responses to resident commensals drives development of pathobiont pathogenicity [135]. These data suggest that expansion of pathobionts in the intestinal mucosa result from the complex interaction of commensals, IECs, and mucosal immune responses all of which contribute to the intestinal microenvironment. The extent of pathobiont-induced dysbiosis can vary as well. For example, Helicobacter bilis does not alter microbial composition while adherent-invasive E. coli induces dysbiosis despite commensal-directed immune responses [135,136]. In this section we review the current understanding of various pathobionts in the GI tract.
Pathobionts can interact with IECs to exert their effects both directly through adherence and translocation and indirectly through modulation of IEC functions (Fig. 1). Pathobionts such as Bilophila wadsworthia and Enterobacteriaceae faecalis are able to adhere to and translocate through IEC monolayers in vitro and in vivo [137-139]. Adherence and invasion of pathobionts usually requires expression of specific genes such as proteases in E. faecalis and GipA (growth in Peyer’s patches) in AIEC [140-143]. In addition to attachment and invasion, pathobiont species also impair normal IEC functions for intestinal homeostasis. C. concisus, a gram-negative microbe that is increased in pediatric and adult IBD patients, inhibits expression of the tight junction-associated proteins ZO-1 and occludin, as well as their association with tight junction complexes leading to a less regulated intestinal barrier [144-147]. Pathobionts can also alter the life cycle of IECs. C. concisus increased apoptosis which left large gaps (apoptotic lesions) in the monolayer that persisted longer than those in control HT-29/B6 IECs [148]. In addition, colibactin-producing E. coli strains (which are also associated with inflammation-associated colorectal cancer) activate senescent secretory intestinal cells to increase growth factor production, leading to increased local cell proliferation and tumor growth [149-151]. Finally, expansion of Bacteroides vulgatus in Nod2-deficient mice was associated with goblet cell dysfunction and subsequently increased numbers of interferon-γ-expressing lymphocytes [152]. Together these studies demonstrated the ability of pathobionts to hijack the proliferation-differentiation process in the intestine in order to reduce important mediators of host defense and promote bacterial invasion.
Pathobiont expansion can also increase pro-inflammatory signaling in IECs leading to dysbiosis and colitis. As mentioned above, B. vulgatus also increased pro-inflammatory cytokine production in Nod2-deficient mice [153]. The capacity of pathobionts to drive a pro-inflammatory response does not appear to be uniformly dependent on the presence of a host defect in innate immunity as E. faecalis activates NF-κB, p38 MAPK, and ERK1/2 pro-inflammatory signaling via TLR2 resulting in induction of IL-6 and IP-10 secretion in wild-type murine IECs [154]. AIEC secretion of outer membrane vesicles leads to activation of the host ER stress response protein Gp96 [155,156]. In addition, AIEC infection in mice also increased Il6 and Lcn2 mRNA expression, fecal lipocalin-2 content, and spleen weight [108]. Bretin and colleagues recently suggested that transient AIEC infection in mice can promote inflammatory signaling after clearance of the pathobiont [136]. These data demonstrate that pathobionts are able to induce stress responses and inflammatory signaling in IECs and may continue to promote inflammation long after infection.
While much work has focused on how pathobionts induce inflammation, we have only begun to understand how these initially innocuous microbes become virulent and expand. As chronic GI disease is prevalent in developed countries, it has been hypothesized that environmental and behavioral factors such as diet contribute to dysbiosis. In support of this, studies have demonstrated that individuals relocating to developed countries begin to exhibit a more Western-like microbiome composition, similar to that of non-human primates who are captured in the wild and transferred to captivity [157]. B. wadsworthia is a gram-negative bacterium usually found at low abundance and has sulfite-reducing activity. Mice that were fed a high milk fat diet with saturated fats developed expanded B. wadsworthia
leading to colitis, however this did not occur in mice fed other types of saturated fats or polyunsaturated fats [117]. Milk fats have elevated levels of hydrophobic stearate that requires higher concentrations of bile salts for emulsification. Milk fat feeding led to a higher ratio of the bile salt taurocholate, which is a more efficient emulsifier and a source of organic sulfur [117]. The subsequent increase in luminal organic sulfur facilitated a bacterial bloom of B. wadsworthia in these mice [117]. In addition to dietary components, inflammatory mediators can also promote pathobiont growth. For example, when IL-22RA1 KO mice were infected with Citrobacter rodentium they developed sepsis and had increased mortality due to expansion and increased translocation of E. faecalis. IL-22RA increases Fut2 expression and subsequently increases intestinal fucosylation. This promotes increased diversity of anaerobic commensal symbionts, restricting expansion of E. faecalis [113]. Pathobionts can also promote their own survival by escaping or resisting host defense mechanisms. For example, E. faecalis can resist antimicrobials targeting its cell envelope through expression of IreK, which is a kinase important for long-term colonization, potentially through inhibition of antagonistic enterococci proteins [158]. Thus, it is clear that a variety of host and environmental factors can precipitate selective expansion of pathobionts and that inflammatory responses can not only alter populations of pathobionts but also modify their level of pathogenicity, in conjunction with increasing host epithelial susceptibility to pathobionts.
It is increasingly clear that the gut microbiota has a significant impact on human health. Recent literature has described how changes in the intestinal microbiota can affect neurological networks and behavior, host metabolism, cardiovascular health, and many other physiological systems. Commensal bacteria promote mucosal health by fortifying barrier integrity, increasing mucin and IgA secretion, inhibiting pro-inflammatory responses and apoptosis, and promoting commensal colonization. In contrast, pathobiont bacteria repress expression of tight junction proteins and promote mislocalization, cause dysregulation of apoptosis and proliferation, and increase ER stress and pro-inflammatory signaling (Fig. 1). These effects contribute to the development of dysbiosis and prolong inflammatory signaling following infection. While much work has begun to unravel this intricate network of interactions between the gut microbiota, IECs, immune cells, and environmental factors, many questions still remain due to limitations in technology and methods to adequately study the microbiome. Future studies focusing on the causal roles of bacteria and inflammation in driving changes to the microbiome and the influence of these changes on the intestinal mucosa will not only increase our understanding of how these relationships operate in different setting (i.e., health vs. disease) but has the potential to identify strategies through which we can harness the therapeutic potential of host-gut microbe interactions.
Notes
REFERENCES
1. Ng SC, Shi HY, Hamidi N, et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet. 2018; 390:2769–2778.

2. Dahlhamer JM, Zammitti EP, Ward BW, Wheaton AG, Croft JB. Prevalence of inflammatory bowel disease among adults aged ≥18 years - United States, 2015. MMWR Morb Mortal Wkly Rep. 2016; 65:1166–1169.

3. Centers for Disease Control and Prevention. Data and statistics: inflammatory bowel disease prevalence (IBD) in the United States. CDC Web site. (www.cdc.gov/ibd/data-statistics.htm. Updated November 14, 2018. Accessed February 13, 2019.
4. Madsen KL, Malfair D, Gray D, Doyle JS, Jewell LD, Fedorak RN. Interleukin-10 gene-deficient mice develop a primary intestinal permeability defect in response to enteric microflora. Inflamm Bowel Dis. 1999; 5:262–270.

5. Roulis M, Bongers G, Armaka M, et al. Host and microbiota interactions are critical for development of murine Crohn’slike ileitis. Mucosal Immunol. 2016; 9:787–797.

6. Khan KJ, Ullman TA, Ford AC, et al. Antibiotic therapy in inflammatory bowel disease: a systematic review and metaanalysis. Am J Gastroenterol. 2011; 106:661–673.

7. Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016; 14:e1002533.

8. Davenport ER, Sanders JG, Song SJ, Amato KR, Clark AG, Knight R. The human microbiome in evolution. BMC Biol. 2017; 15:127.

9. Maier E, Anderson RC, Roy NC. Understanding how commensal obligate anaerobic bacteria regulate immune functions in the large intestine. Nutrients. 2014; 7:45–73.

10. Omenetti S, Pizarro TT. The Treg/Th17 axis: a dynamic balance regulated by the gut microbiome. Front Immunol. 2015; 6:639.

11. Lee N, Kim WU. Microbiota in T-cell homeostasis and inflammatory diseases. Exp Mol Med. 2017; 49:e340.

12. Verma R, Lee C, Jeun EJ, et al. Cell surface polysaccharides of Bifidobacterium bifidum induce the generation of Foxp3(+) regulatory T cells. Sci Immunol. 2018; 3:eaat6975.

13. Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006; 444:1027–1031.

14. Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007; 104:13780–13785.

15. Levy M, Kolodziejczyk AA, Thaiss CA, Elinav E. Dysbiosis and the immune system. Nat Rev Immunol. 2017; 17:219–232.

16. Brown EM, Sadarangani M, Finlay BB. The role of the immune system in governing host-microbe interactions in the intestine. Nat Immunol. 2013; 14:660–667.

17. Watson AJ, Duckworth CA, Guan Y, Montrose MH. Mechanisms of epithelial cell shedding in the mammalian intestine and maintenance of barrier function. Ann N Y Acad Sci. 2009; 1165:135–142.

18. Marchiando AM, Shen L, Graham WV, et al. The epithelial barrier is maintained by in vivo tight junction expansion during pathologic intestinal epithelial shedding. Gastroenterology. 2011; 140:1208–1218.

19. Blander JM. Death in the intestinal epithelium-basic biology and implications for inflammatory bowel disease. FEBS J. 2016; 283:2720–2730.

20. Tsai PY, Zhang B, He WQ, et al. IL-22 upregulates epithelial claudin-2 to drive diarrhea and enteric pathogen clearance. Cell Host Microbe. 2017; 21:671–681.
21. Chairatana P, Nolan EM. Defensins, lectins, mucins, and secretory immunoglobulin A: microbe-binding biomolecules that contribute to mucosal immunity in the human gut. Crit Rev Biochem Mol Biol. 2017; 52:45–56.

22. Ermund A, Schütte A, Johansson ME, Gustafsson JK, Hansson GC. Studies of mucus in mouse stomach, small intestine, and colon. I. Gastrointestinal mucus layers have different properties depending on location as well as over the Peyer’s patches. Am J Physiol Gastrointest Liver Physiol. 2013; 305:G341–G347.

23. Johansson ME, Jakobsson HE, Holmén-Larsson J, et al. Normalization of host intestinal mucus layers requires long-term microbial colonization. Cell Host Microbe. 2015; 18:582–592.

25. Norkina O, Burnett TG, De Lisle RC. Bacterial overgrowth in the cystic fibrosis transmembrane conductance regulator null mouse small intestine. Infect Immun. 2004; 72:6040–6049.

26. Bazett M, Honeyman L, Stefanov AN, Pope CE, Hoffman LR, Haston CK. Cystic fibrosis mouse model-dependent intestinal structure and gut microbiome. Mamm Genome. 2015; 26:222–234.

27. Price AE, Shamardani K, Lugo KA, et al. A map of Toll-like receptor expression in the intestinal epithelium reveals distinct spatial, cell type-specific, and temporal patterns. Immunity. 2018; 49:560–575.

28. Vora P, Youdim A, Thomas LS, et al. Beta-defensin-2 expression is regulated by TLR signaling in intestinal epithelial cells. J Immunol. 2004; 173:5398–5405.

29. Cario E, Gerken G, Podolsky DK. Toll-like receptor 2 controls mucosal inflammation by regulating epithelial barrier function. Gastroenterology. 2007; 132:1359–1374.

30. Ulluwishewa D, Anderson RC, McNabb WC, Moughan PJ, Wells JM, Roy NC. Regulation of tight junction permeability by intestinal bacteria and dietary components. J Nutr. 2011; 141:769–776.

31. Ivanov II, Atarashi K, Manel N, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009; 139:485–498.

32. Goto Y, Panea C, Nakato G, et al. Segmented filamentous bacteria antigens presented by intestinal dendritic cells drive mucosal Th17 cell differentiation. Immunity. 2014; 40:594–607.

33. Atarashi K, Tanoue T, Ando M, et al. Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell. 2015; 163:367–380.

34. Sano T, Huang W, Hall JA, et al. An IL-23R/IL-22 circuit regulates epithelial serum amyloid a to promote local effector Th17 responses. Cell. 2015; 163:381–393.

35. Tan TG, Sefik E, Geva-Zatorsky N, et al. Identifying species of symbiont bacteria from the human gut that, alone, can induce intestinal Th17 cells in mice. Proc Natl Acad Sci U S A. 2016; 113:E8141–E8150.

36. Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005; 122:107–118.

37. Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008; 453:620–625.

38. Round JL, Mazmanian SK. Inducible Foxp3+ regulatory Tcell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A. 2010; 107:12204–12209.

39. Knoop KA, Newberry RD. Isolated lymphoid follicles are dynamic reservoirs for the induction of intestinal IgA. Front Immunol. 2012; 3:84.

40. Wu W, Sun M, Chen F, et al. Microbiota metabolite short-chain fatty acid acetate promotes intestinal IgA response to microbiota which is mediated by GPR43. Mucosal Immunol. 2017; 10:946–956.

41. Wang Y, Liu L, Moore DJ, et al. An LGG-derived protein promotes IgA production through upregulation of APRIL expression in intestinal epithelial cells. Mucosal Immunol. 2017; 10:373–384.

42. Bollinger RR, Everett ML, Palestrant D, Love SD, Lin SS, Parker W. Human secretory immunoglobulin A may contribute to biofilm formation in the gut. Immunology. 2003; 109:580–587.

43. Mathias A, Duc M, Favre L, Benyacoub J, Blum S, Corthésy B. Potentiation of polarized intestinal Caco-2 cell responsiveness to probiotics complexed with secretory IgA. J Biol Chem. 2010; 285:33906–33913.

44. Donaldson GP, Ladinsky MS, Yu KB, et al. Gut microbiota utilize immunoglobulin A for mucosal colonization. Science. 2018; 360:795–800.

45. Palm NW, de Zoete MR, Cullen TW, et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. 2014; 158:1000–1010.

46. Hugot JP, Chamaillard M, Zouali H, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001; 411:599–603.

47. Barrett JC, Hansoul S, Nicolae DL, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet. 2008; 40:955–962.
48. Franke A, McGovern DP, Barrett JC, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010; 42:1118–1125.
49. McGovern DP, Gardet A, Törkvist L, et al. Genome-wide association identifies multiple ulcerative colitis susceptibility loci. Nat Genet. 2010; 42:332–337.
50. Anderson CA, Boucher G, Lees CW, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011; 43:246–252.
51. Petnicki-Ocwieja T, Hrncir T, Liu YJ, et al. Nod2 is required for the regulation of commensal microbiota in the intestine. Proc Natl Acad Sci U S A. 2009; 106:15813–15818.

52. Rehman A, Sina C, Gavrilova O, et al. Nod2 is essential for temporal development of intestinal microbial communities. Gut. 2011; 60:1354–1362.

53. Mondot S, Barreau F, Al Nabhani Z, et al. Altered gut microbiota composition in immune-impaired Nod2(-/-) mice. Gut. 2012; 61:634–635.

54. Sadaghian Sadabad M, Regeling A, de Goffau MC, et al. The ATG16L1-T300A allele impairs clearance of pathosymbionts in the inflamed ileal mucosa of Crohn’s disease patients. Gut. 2015; 64:1546–1552.

55. Pott J, Kabat AM, Maloy KJ. Intestinal epithelial cell autophagy is required to protect against TNF-induced apoptosis during chronic colitis in mice. Cell Host Microbe. 2018; 23:191–202.

56. McCole DF. IBD candidate genes and intestinal barrier regulation. Inflamm Bowel Dis. 2014; 20:1829–1849.

57. Knights D, Silverberg MS, Weersma RK, et al. Complex host genetics influence the microbiome in inflammatory bowel disease. Genome Med. 2014; 6:107.

58. Ng KM, Ferreyra JA, Higginbottom SK, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013; 502:96–99.

59. Xu J, Bjursell MK, Himrod J, et al. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science. 2003; 299:2074–2076.

60. Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucindegrading bacterium. Int J Syst Evol Microbiol. 2004; 54:1469–1476.

61. Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005; 307:1915–1920.

62. Png CW, Lindén SK, Gilshenan KS, et al. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol. 2010; 105:2420–2428.

63. van Passel MW, Kant R, Zoetendal EG, et al. The genome of Akkermansia muciniphila, a dedicated intestinal mucin degrader, and its use in exploring intestinal metagenomes. PLoS One. 2011; 6:e16876.

64. Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol. 2010; 10:159–169.

65. Van den Abbeele P, Belzer C, Goossens M, et al. Butyrate-producing Clostridium cluster XIVa species specifically colonize mucins in an in vitro gut model. ISME J. 2013; 7:949–961.

66. Robinson LS, Lewis WG, Lewis AL. The sialate O-acetylesterase EstA from gut Bacteroidetes species enables sialidasemediated cross-species foraging of 9-O-acetylated sialoglycans. J Biol Chem. 2017; 292:11861–11872.

67. Lopez-Siles M, Khan TM, Duncan SH, Harmsen HJ, Garcia-Gil LJ, Flint HJ. Cultured representatives of two major phylogroups of human colonic Faecalibacterium prausnitzii can utilize pectin, uronic acids, and host-derived substrates for growth. Appl Environ Microbiol. 2012; 78:420–428.

68. Belzer C, Chia LW, Aalvink S, et al. Microbial metabolic networks at the mucus layer lead to diet-independent butyrate and vitamin B(12) production by intestinal symbionts. MBio. 2017; 85:e00770–17.
69. Berry D, Stecher B, Schintlmeister A, et al. Host-compound foraging by intestinal microbiota revealed by single-cell stable isotope probing. Proc Natl Acad Sci U S A. 2013; 110:4720–4725.

70. Reunanen J, Kainulainen V, Huuskonen L, et al. Akkermansia muciniphila adheres to enterocytes and strengthens the integrity of the epithelial cell layer. Appl Environ Microbiol. 2015; 81:3655–3662.

71. Ouwerkerk JP, van der Ark KCH, Davids M, et al. Adaptation of Akkermansia muciniphila to the oxic-anoxic interface of the mucus layer. Appl Environ Microbiol. 2016; 82:6983–6993.

72. Everard A, Belzer C, Geurts L, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A. 2013; 110:9066–9071.

73. Shin NR, Lee JC, Lee HY, et al. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut. 2014; 63:727–735.

74. Ottman N, Reunanen J, Meijerink M, et al. Pili-like proteins of Akkermansia muciniphila modulate host immune responses and gut barrier function. PLoS One. 2017; 12:e0173004.

75. Gaudier E, Jarry A, Blottière HM, et al. Butyrate specifically modulates MUC gene expression in intestinal epithelial goblet cells deprived of glucose. Am J Physiol Gastrointest Liver Physiol. 2004; 287:G1168–G1174.
76. Thibault R, Blachier F, Darcy-Vrillon B, de Coppet P, Bourreille A, Segain JP. Butyrate utilization by the colonic mucosa in inflammatory bowel diseases: a transport deficiency. Inflamm Bowel Dis. 2010; 16:684–695.

77. Koh A, De Vadder F, Kovatcheva-Datchary P, Bäckhed F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell. 2016; 165:1332–1345.

78. Litvak Y, Byndloss MX, Bäumler AJ. Colonocyte metabolism shapes the gut microbiota. Science. 2018; 362:eaat9076.

79. Alam A, Leoni G, Quiros M, et al. The microenvironment of injured murine gut elicits a local pro-restitutive microbiota. Nat Microbiol. 2016; 1:15021.

80. Rajilić-Stojanović M, Shanahan F, Guarner F, de Vos WM. Phylogenetic analysis of dysbiosis in ulcerative colitis during remission. Inflamm Bowel Dis. 2013; 19:481–488.

81. Swidsinski A, Dörffel Y, Loening-Baucke V, et al. Acute appendicitis is characterised by local invasion with Fusobacterium nucleatum/necrophorum. Gut. 2011; 60:34–40.

82. Karlsson CL, Onnerfält J, Xu J, Molin G, Ahrné S, Thorngren-Jerneck K. The microbiota of the gut in preschool children with normal and excessive body weight. Obesity (Silver Spring). 2012; 20:2257–2261.

83. Dao MC, Everard A, Aron-Wisnewsky J, et al. Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: relationship with gut microbiome richness and ecology. Gut. 2016; 65:426–436.

84. Zhang YG, Wu S, Xia Y, Sun J. Salmonella infection upregulates the leaky protein claudin-2 in intestinal epithelial cells. PLoS One. 2013; 8:e58606.

85. Chelakkot C, Choi Y, Kim DK, et al. Akkermansia muciniphila-derived extracellular vesicles influence gut permeability through the regulation of tight junctions. Exp Mol Med. 2018; 50:e450.

86. Wlodarska M, Luo C, Kolde R, et al. Indoleacrylic acid produced by commensal peptostreptococcus species suppresses inflammation. Cell Host Microbe. 2017; 22:25–37.

87. Sokol H, Seksik P, Furet JP, et al. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis. 2009; 15:1183–1189.

88. Lopez-Siles M, Martinez-Medina M, Abellà C, et al. Mucosa-associated Faecalibacterium prausnitzii phylotype richness is reduced in patients with inflammatory bowel disease. Appl Environ Microbiol. 2015; 81:7582–7592.

89. Lopez-Siles M, Enrich-Capó N, Aldeguer X, et al. Alterations in the abundance and co-occurrence of Akkermansia muciniphila and Faecalibacterium prausnitzii in the colonic mucosa of inflammatory bowel disease subjects. Front Cell Infect Microbiol. 2018; 8:281.

90. Arumugam M, Raes J, Pelletier E, et al. Enterotypes of the human gut microbiome. Nature. 2011; 473:174–180.

91. Cao Y, Shen J, Ran ZH. Association between Faecalibacterium prausnitzii reduction and inflammatory bowel disease: a meta-analysis and systematic review of the literature. Gastroenterol Res Pract. 2014; 2014:872725.
92. Sokol H, Pigneur B, Watterlot L, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008; 105:16731–16736.

93. Jia W, Whitehead RN, Griffiths L, et al. Is the abundance of Faecalibacterium prausnitzii relevant to Crohn’s disease? FEMS Microbiol Lett. 2010; 310:138–144.

94. Quévrain E, Maubert MA, Michon C, et al. Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn’s disease. Gut. 2016; 65:415–425.

95. Breyner NM, Michon C, de Sousa CS, et al. Microbial anti-inflammatory molecule (MAM) from Faecalibacterium prausnitzii shows a protective effect on DNBS and DSS-induced colitis model in mice through inhibition of NF-kappaB pathway. Front Microbiol. 2017; 8:114.
96. Maier E, Anderson RC, Roy NC. Live Faecalibacterium prausnitzii does not enhance epithelial barrier integrity in an apical anaerobic co-culture model of the large intestine. Nutrients. 2017; 9:1349.

97. He X, Zeng Q, Puthiyakunnon S, et al. Lactobacillus rhamnosus GG supernatant enhance neonatal resistance to systemic Escherichia coli K1 infection by accelerating development of intestinal defense. Sci Rep. 2017; 7:43305.

98. Alam A, Leoni G, Wentworth CC, et al. Redox signaling regulates commensal-mediated mucosal homeostasis and restitution and requires formyl peptide receptor 1. Mucosal Immunol. 2014; 7:645–655.

99. Yan F, Cao H, Cover TL, Whitehead R, Washington MK, Polk DB. Soluble proteins produced by probiotic bacteria regulate intestinal epithelial cell survival and growth. Gastroenterology. 2007; 132:562–575.

100. Shen X, Liu L, Peek RM, et al. Supplementation of p40, a Lactobacillus rhamnosus GG-derived protein, in early life promotes epidermal growth factor receptor-dependent intestinal development and long-term health outcomes. Mucosal Immunol. 2018; 11:1316–1328.

101. Baughn AD, Malamy MH. The strict anaerobe Bacteroides fragilis grows in and benefits from nanomolar concentrations of oxygen. Nature. 2004; 427:441–444.

102. Tajkarimi M, Wexler HM. CRISPR-Cas systems in Bacteroides fragilis, an important pathobiont in the human gut microbiome. Front Microbiol. 2017; 8:2234.

103. Chan JL, Wu S, Geis AL, et al. Non-toxigenic Bacteroides fragilis (NTBF) administration reduces bacteria-driven chronic colitis and tumor development independent of polysaccharide A. Mucosal Immunol. 2019; 12:164–177.

104. Scanlan PD, Shanahan F, O’Mahony C, Marchesi JR. Culture-independent analyses of temporal variation of the dominant fecal microbiota and targeted bacterial subgroups in Crohn’s disease. J Clin Microbiol. 2006; 44:3980–3988.

105. Patwa LG, Fan TJ, Tchaptchet S, et al. Chronic intestinal inflammation induces stress-response genes in commensal Escherichia coli. Gastroenterology. 2011; 141:1842–1851.

106. Tchaptchet S, Fan TJ, Goeser L, et al. Inflammation-induced acid tolerance genes gadAB in luminal commensal Escherichia coli attenuate experimental colitis. Infect Immun. 2013; 81:3662–3671.

107. Ocvirk S, Sava IG, Lengfelder I, et al. Surface-associated lipoproteins link Enterococcus faecalis virulence to colitogenic activity in IL-10-deficient mice independent of their expression levels. PLoS Pathog. 2015; 11:e1004911.

108. Chassaing B, Gewirtz AT. Mice harboring pathobiont-free microbiota do not develop intestinal inflammation that normally results from an innate immune deficiency. PLoS One. 2018; 13:e0195310.

109. Haberman Y, Tickle TL, Dexheimer PJ, et al. Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature. J Clin Invest. 2014; 124:3617–3633.

110. Swidsinski A, Loening-Baucke V, Verstraelen H, Osowska S, Doerffel Y. Biostructure of fecal microbiota in healthy subjects and patients with chronic idiopathic diarrhea. Gastroenterology. 2008; 135:568–579.

111. Medani M, Collins D, Docherty NG, Baird AW, O’Connell PR, Winter DC. Emerging role of hydrogen sulfide in colonic physiology and pathophysiology. Inflamm Bowel Dis. 2011; 17:1620–1625.

112. Thiennimitr P, Winter SE, Winter MG, et al. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci U S A. 2011; 108:17480–17485.

113. Pham TA, Clare S, Goulding D, et al. Epithelial IL-22RA1-mediated fucosylation promotes intestinal colonization resistance to an opportunistic pathogen. Cell Host Microbe. 2014; 16:504–516.

114. Zhu W, Winter MG, Byndloss MX, et al. Precision editing of the gut microbiota ameliorates colitis. Nature. 2018; 553:208–211.

115. Deriu E, Liu JZ, Pezeshki M, et al. Probiotic bacteria reduce Salmonella typhimurium intestinal colonization by competing for iron. Cell Host Microbe. 2013; 14:26–37.

116. Bolick DT, Kolling GL, Moore JH 2nd, et al. Zinc deficiency alters host response and pathogen virulence in a mouse model of enteroaggregative Escherichia coli-induced diarrhea. Gut Microbes. 2014; 5:618–627.

117. Devkota S, Wang Y, Musch MW, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10-/- mice. Nature. 2012; 487:104–108.

118. Franzosa EA, Sirota-Madi A, Avila-Pacheco J, et al. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat Microbiol. 2019; 4:293–305.

119. Albenberg L, Esipova TV, Judge CP, et al. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology. 2014; 147:1055–1063.

120. Lupp C, Robertson ML, Wickham ME, et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007; 2:204.

121. Rivera-Chávez F, Zhang LF, Faber F, et al. Depletion of butyrate-producing clostridia from the gut microbiota drives an aerobic luminal expansion of Salmonella. Cell Host Microbe. 2016; 19:443–454.

122. Fiorucci S, Distrutti E, Cirino G, Wallace JL. The emerging roles of hydrogen sulfide in the gastrointestinal tract and liver. Gastroenterology. 2006; 131:259–271.

123. Bertin Y, Girardeau JP, Chaucheyras-Durand F, et al. Enterohaemorrhagic Escherichia coli gains a competitive advantage by using ethanolamine as a nitrogen source in the bovine intestinal content. Environ Microbiol. 2011; 13:365–377.

124. Faber F, Thiennimitr P, Spiga L, et al. Respiration of microbiota-derived 1,2-propanediol zdrives Salmonella expansion during colitis. PLoS Pathog. 2017; 13:e1006129.
125. Maslowski KM, Vieira AT, Ng A, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009; 461:1282–1286.

126. Hughes ER, Winter MG, Duerkop BA, et al. Microbial respiration and formate oxidation as metabolic signatures of inflammation-associated dysbiosis. Cell Host Microbe. 2017; 21:208–219.

127. Lopez CA, Skaar EP. The impact of dietary transition metals on host-bacterial interactions. Cell Host Microbe. 2018; 23:737–748.

128. Blachier F, Beaumont M, Andriamihaja M, et al. Changes in the luminal environment of the colonic epithelial cells and physiopathological consequences. Am J Pathol. 2017; 187:476–486.

129. Walker AW, Duncan SH, McWilliam Leitch EC, Child MW, Flint HJ. pH and peptide supply can radically alter bacterial populations and short-chain fatty acid ratios within microbial communities from the human colon. Appl Environ Microbiol. 2005; 71:3692–3700.

130. Talbot C, Lytle C. Segregation of Na/H exchanger-3 and Cl/HCO3 exchanger SLC26A3 (DRA) in rodent cecum and colon. Am J Physiol Gastrointest Liver Physiol. 2010; 299:G358–G367.
131. Vernia P, Caprilli R, Latella G, Barbetti F, Magliocca FM, Cittadini M. Fecal lactate and ulcerative colitis. Gastroenterology. 1988; 95:1564–1568.

132. Belenguer A, Duncan SH, Holtrop G, Anderson SE, Lobley GE, Flint HJ. Impact of pH on lactate formation and utilization by human fecal microbial communities. Appl Environ Microbiol. 2007; 73:6526–6533.

133. Chow J, Mazmanian SK. A pathobiont of the microbiota balances host colonization and intestinal inflammation. Cell Host Microbe. 2010; 7:265–276.

134. Wu C, Sartor RB, Huang K, Tonkonogy SL. Transient activation of mucosal effector immune responses by resident intestinal bacteria in normal hosts is regulated by interleukin-10 signalling. Immunology. 2016; 148:304–314.

135. Gomes-Neto JC, Kittana H, Mantz S, et al. A gut pathobiont synergizes with the microbiota to instigate inflammatory disease marked by immunoreactivity against other symbionts but not itself. Sci Rep. 2017; 7:17707.

136. Bretin A, Lucas C, Larabi A, et al. AIEC infection triggers modification of gut microbiota composition in genetically predisposed mice, contributing to intestinal inflammation. Sci Rep. 2018; 8:12301.

137. Wells CL, Jechorek RP, Erlandsen SL. Evidence for the translocation of Enterococcus faecalis across the mouse intestinal tract. J Infect Dis. 1990; 162:82–90.

138. Runkel NS, Moody FG, Smith GS, et al. Alterations in rat intestinal transit by morphine promote bacterial translocation. Dig Dis Sci. 1993; 38:1530–1536.

139. Vinderola CG, Medici M, Perdigón G. Relationship between interaction sites in the gut, hydrophobicity, mucosal immunomodulating capacities and cell wall protein profiles in indigenous and exogenous bacteria. J Appl Microbiol. 2004; 96:230–243.

140. Gerardo SH, Garcia MM, Wexler HM, Finegold SM. Adherence of Bilophila wadsworthia to cultured human embryonic intestinal cells. Anaerobe. 1998; 4:19–27.

141. Zeng J, Teng F, Murray BE. Gelatinase is important for translocation of Enterococcus faecalis across polarized human enterocyte-like T84 cells. Infect Immun. 2005; 73:1606–1612.

142. Zeng J, Teng F, Weinstock GM, Murray BE. Translocation of Enterococcus faecalis strains across a monolayer of polarized human enterocyte-like T84 cells. J Clin Microbiol. 2004; 42:1149–1154.

143. Vazeille E, Chassaing B, Buisson A, et al. GipA factor supports colonization of Peyer’s patches by Crohn’s disease-associated Escherichia coli. Inflamm Bowel Dis. 2016; 22:68–81.

144. Zhang L, Man SM, Day AS, et al. Detection and isolation of Campylobacter species other than C. jejuni from children with Crohn’s disease. J Clin Microbiol. 2009; 47:453–455.

145. Mahendran V, Riordan SM, Grimm MC, et al. Prevalence of Campylobacter species in adult Crohn’s disease and the preferential colonization sites of Campylobacter species in the human intestine. PLoS One. 2011; 6:e25417.

146. Mukhopadhya I, Thomson JM, Hansen R, Berry SH, El-Omar EM, Hold GL. Detection of Campylobacter concisus and other Campylobacter species in colonic biopsies from adults with ulcerative colitis. PLoS One. 2011; 6:e21490.

147. Man SM, Kaakoush NO, Leach ST, et al. Host attachment, invasion, and stimulation of proinflammatory cytokines by Campylobacter concisus and other non-Campylobacter jejuni Campylobacter species. J Infect Dis. 2010; 202:1855–1865.

148. Nielsen HL, Nielsen H, Ejlertsen T, et al. Oral and fecal Campylobacter concisus strains perturb barrier function by apoptosis induction in HT-29/B6 intestinal epithelial cells. PLoS One. 2011; 6:e23858.

149. Raisch J, Buc E, Bonnet M, et al. Colon cancer-associated B2 Escherichia coli colonize gut mucosa and promote cell proliferation. World J Gastroenterol. 2014; 20:6560–6572.

150. Cougnoux A, Dalmasso G, Martinez R, et al. Bacterial genotoxin colibactin promotes colon tumour growth by inducing a senescence-associated secretory phenotype. Gut. 2014; 63:1932–1942.

151. Arthur JC, Jobin C. The complex interplay between inflammation, the microbiota and colorectal cancer. Gut Microbes. 2013; 4:253–258.
152. Ramanan D, Tang MS, Bowcutt R, Loke P, Cadwell K. Bacterial sensor Nod2 prevents inflammation of the small intestine by restricting the expansion of the commensal Bacteroides vulgatus. Immunity. 2014; 41:311–324.

153. Charlet R, Pruvost Y, Tumba G, et al. Remodeling of the Candida glabrata cell wall in the gastrointestinal tract affects the gut microbiota and the immune response. Sci Rep. 2018; 8:3316.

154. Hoffmann M, Messlik A, Kim SC, Sartor RB, Haller D. Impact of a probiotic Enterococcus faecalis in a gnotobiotic mouse model of experimental colitis. Mol Nutr Food Res. 2011; 55:703–713.

155. Rolhion N, Barnich N, Bringer MA, et al. Abnormally expressed ER stress response chaperone Gp96 in CD favours adherent-invasive Escherichia coli invasion. Gut. 2010; 59:1355–1362.

156. Rolhion N, Hofman P, Darfeuille-Michaud A. The endoplasmic reticulum stress response chaperone: Gp96, a host receptor for Crohn disease-associated adherent-invasive Escherichia coli. Gut Microbes. 2011; 2:115–119.

Fig. 1.
Effect of bacteria on intestinal epithelial cells (IECs) leading to mucosal health or disease. Symbionts/commensal bacteria facilitate intestinal health by promoting tolerogenic immunity and barrier function. Expansion of pathobionts can lead to alteration of epithelial turnover, increased endoplasmic reticulum (ER) stress, pro-inflammatory signaling, and impaired barrier function. TJ, tight junction; Treg, T regulatory cell; AMP, antimicrobial peptide.
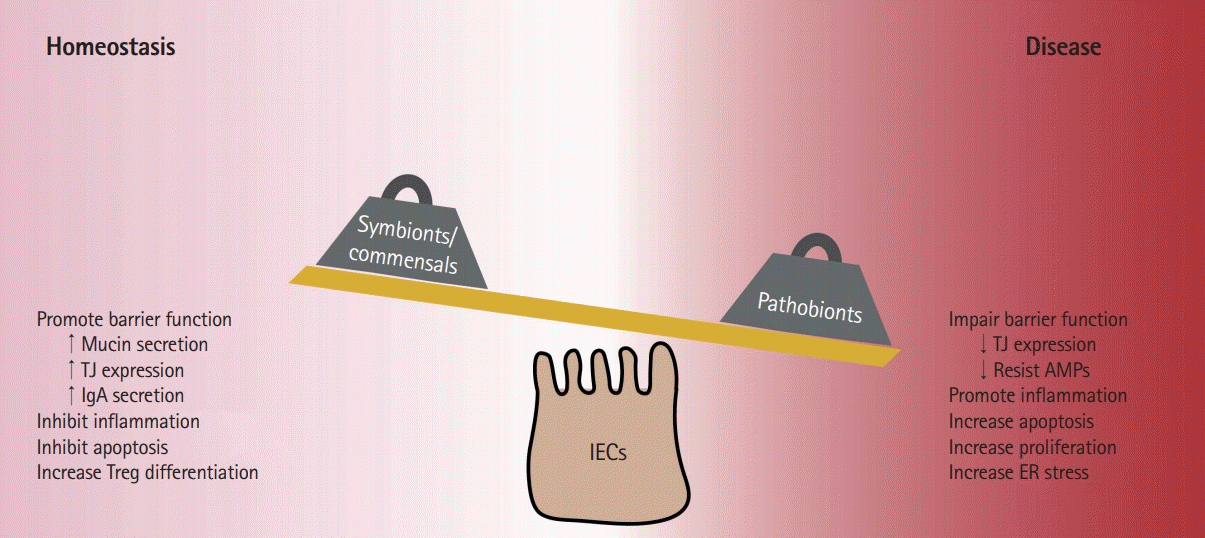
Table 1.
Factors Influencing Microbial Colonization
| Factor | Regulated by | Effect |
|---|---|---|
| O2 concentration | Butyrate producers decrease O2 (antibiotic therapy) | Decreased numbers of commensal microbes and increased pathogenic bacteria [109] |
| Inflammation (ROS secretion and increased blood flow) increase O2 | Increased number of aerotolerant bacteria [110] | |
| Decreased at sites of injury | Akkermansia muciniphila increases mucosal healing wound sites [79] | |
| Reactive oxygen/nitrogen species | Inflammation | Increase O2 concentration [110] |
| Promote alternative electron acceptors for microbial respiration, increasing pathogen number [78,111] | ||
| Inflammatory mediators | Inflammation–ethanolamine | Promotes expansion of pathogenic bacteria [112] |
| Inflammation–IL22RA1 | Increases intestinal fucosylation promoting diversity of anaerobic commensals and represses Enterococcus faecalis [113] | |
| Dietary transition metals | Inflammation–molybdenum | Utilized for microbial respiration to promote Enterobacteriaceae expansion [114] |
| Inflammation–iron | Utilization by Escherichia coli Nissle 1917 restricts expansion of pathogenic bacteria [115] | |
| Zinc | Deficiency increases enteroaggregative E. coli virulence [116] | |
| Dietary metabolites | Host behavior and environment | Increased sulfur promotes Bilophila wadsworthia expansion [117] |
| Milk fats | ||
| pH gradients | Substrate fermentation | Shifts in microbial communities [118] |




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download