Introduction
Materials and Methods
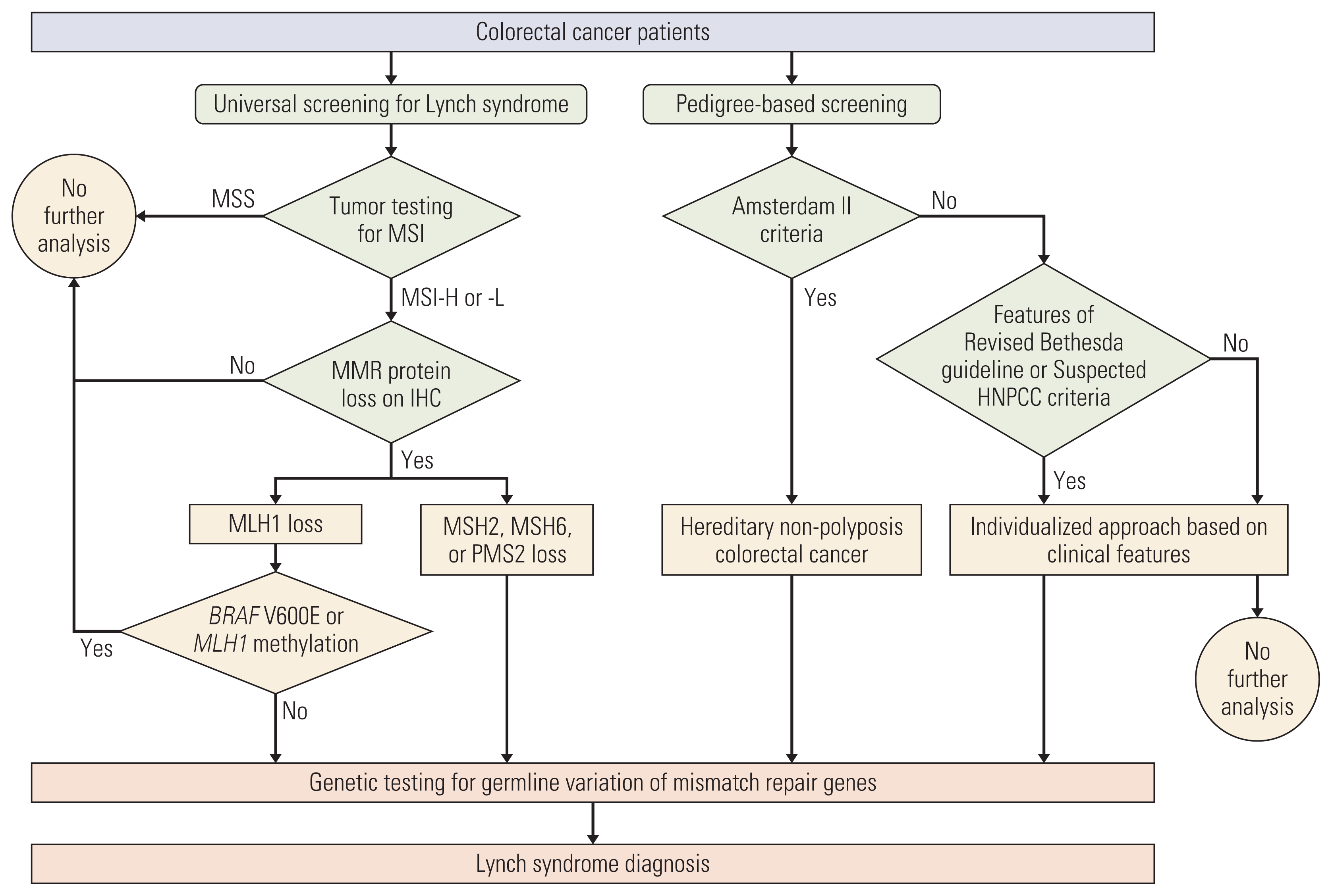
1. Patients
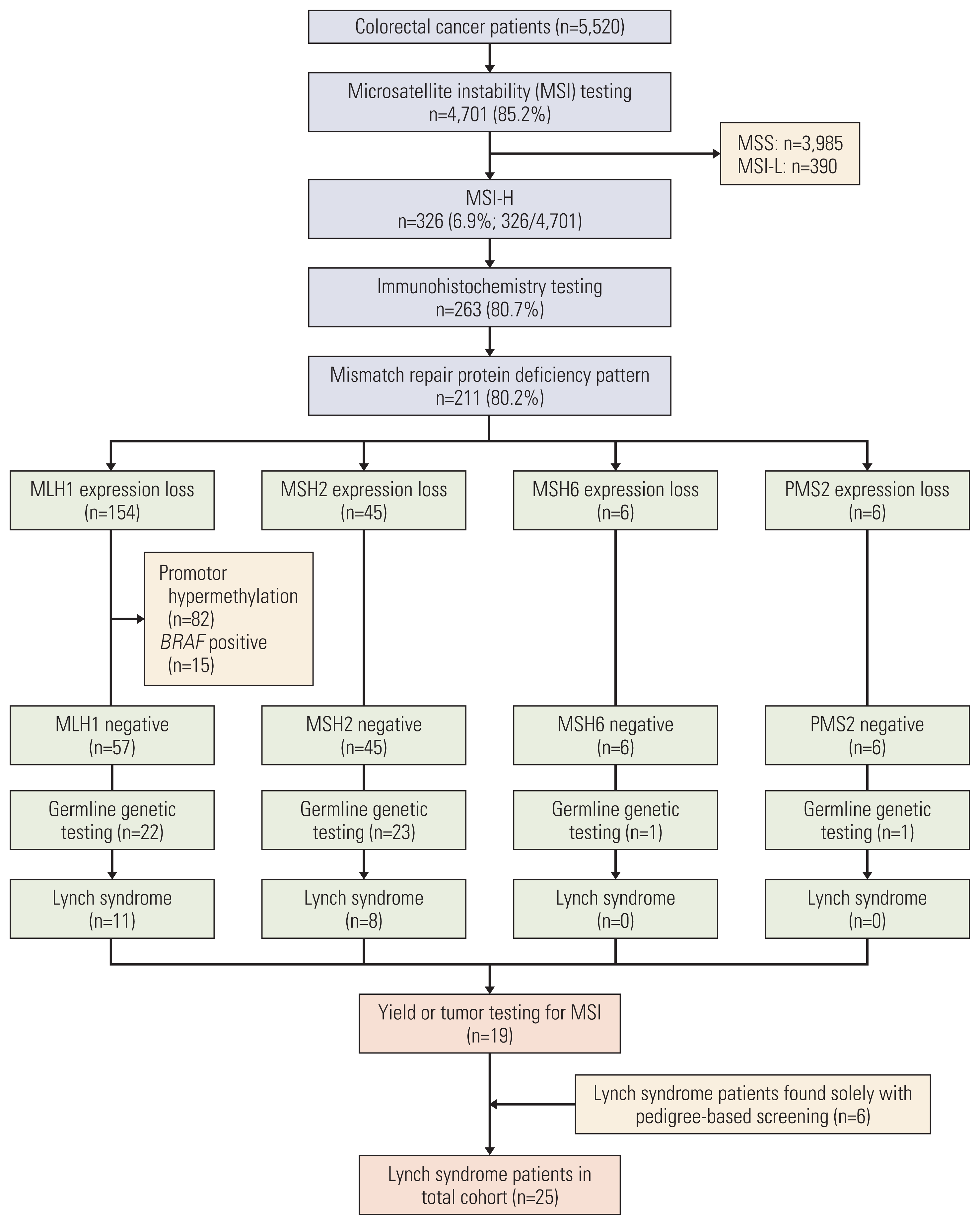
2. Tumor testing for MSI, IHC, and germline genetic testing
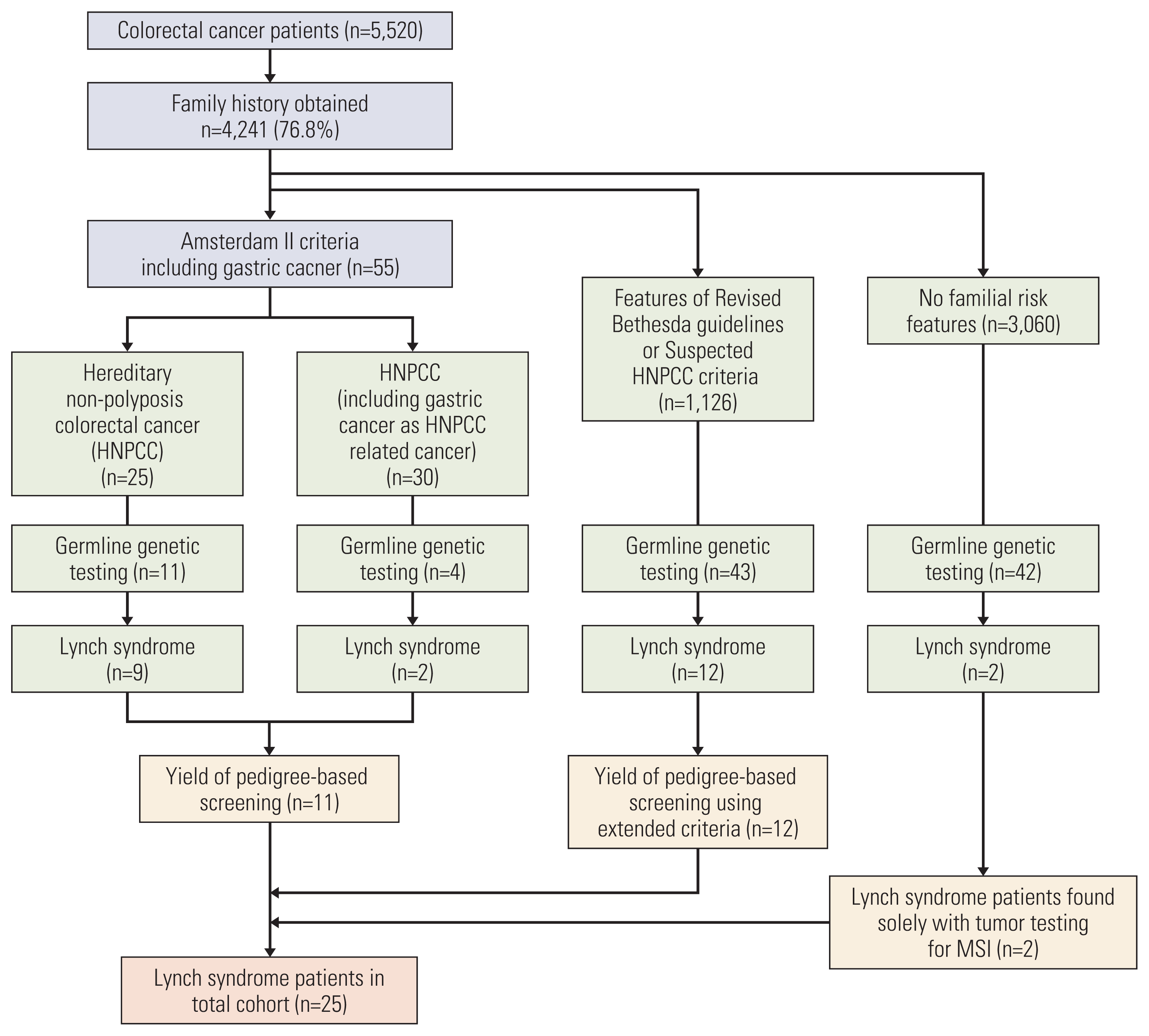
3. Obtaining pedigree and diagnosis of HNPCC
4. Yield analysis
Table 1
| Guideline | Guideline criteria |
|---|---|
| Amsterdam II criteria [16] | There should be at least three relatives with and HNPCC-associated cancer. One should be a first-degree relative of the other two. At least two successive generations should be affected. At least one should be diagnosed before age 50. |
| Revised Bethesda guidelines [18] |
Tumors from individuals should be tested for MSI in the following situations:
|
| Suspected HNPCC criteria [19] | At least two HNPCC-associated cancers in first-degree relatives and |
Results
1. Patients
Table 2
| No. | Sex/Age | Synchronous/Metachronous cancers, age of diagnosis (yr) | Tumor location | Differentiation | Tumor stage | MSI | IHC | MMR pattern | Mutation gene | Grouped by pedigree analysis | Diagnosed with universal screening | PREMM5 prediction score (%)a) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | F/29 | Endometrial, 29 | Rectum | WD | 0 | MSI-H | +/nd/nd/nd | Insufficient | MSH2 c.256G>T | HNPCC | No | > 50 |
| 2 | M/40 | Duodenal, 40 | A-colon | WD | 1 | MSI-H | +/−/−/+ | MSH2 | MSH2 c.687dupA | HNPCC | Yes | 44.4 |
| 3 | F/34 | - | A-colon | MD | 2 | MSI-H | +/−/−/+ | MSH2 | MSH2 c.402delT | Suspicious | Yes | 14.6 |
| 4 | M/41 | - | S-colon | MD | 3 | MSI-H | −/+/+/− | MLH1 | MLH1 c.1409+1G>A | Suspicious | Yes | 11.4 |
| 5 | F/37 | - | A-colon | PD | 2 | MSI-H | −/+/+/− | MLH1 | MLH1 c.1039-4_1068del | Suspicious | Yes | 18.3 |
| 6 | F/55 | Gastric, 61 | Multiple | WD | 2 | MSI-H | −/+/+/− | MLH1 | MLH1 c.790+2T>A | Suspicious | Yes | 15.4 |
| 7 | F/44 | - | Multiple | MD | 3 | MSI-H | −/+/+/− | MLH1 | MLH1 c.1409+1G>A | Suspicious | Yes | 5.9 |
| 8 | M/56 | Small bowel sarcoma, 56 | S-colon | WD | 2 | MSI-H | +/+/nd/nd | Insufficient | MLH1 c.1772_1775delATAG | HNPCC | No | 29.9 |
| 9 | F/29 | - | A-colon | MD | 2 | MSI-H | −/+/+/− | MLH1 | MLH1 c.1667+1G>A | HNPCC | Yes | > 50 |
| 10 | F/40 | - | A-colon | MD | 3 | MSI-H | −/+/+/− | MLH1 | MLH1 c.67G>T | Suspicious | Yes | 4.7 |
| 11 | F/49 | T-colon, 52 | A-colon | PD | 2 | MSI-H | +/−/−/+ | MSH2 | MSH2 c.226C>T | Suspicious | Yes | 3.4 |
| 12 | M/50 | - | T-colon | MD | 2 | MSI-H | −/+/+/− | MLH1 | MLH1 c.67G>T | HNPCC | Yes | 30.4 |
| 13 | M/61 | Thyroid, 64 | A-colon | MD | 1 | MSI-H | +/−/−/+ | MSH2 | MSH2 c.1147c>T | Suspicious | Yes | 3.6 |
| 14 | F/61 | Endometrial, 63 | A-colon | WD | 1 | MSS | +/+/+/+ | All positive | MSH2 c.1580delG | Suspicious | No | > 50 |
| 15 | M/52 | - | A-colon | MD | 2 | MSS | +/−/−/+ | MSH2 | MSH2 c.2699C>A | HNPCC | No | > 50 |
| 16 | M/46 | - | A-colon | MD | 2 | MSI-H | +/−/−/+ | MSH2 | MSH2 c.256G>T | HNPCC | Yes | > 50 |
| 17 | M/47 | - | Rectum | WD | 2 | MSI-H | +/−/−/+ | MSH2 | MSH2 c.2089T>C | HNPCC | Yes | 39.7 |
| 18 | F/47 | - | A-colon | MD | 2 | MSI-H | −/+/+/− | MLH1 | MLH1 c.1721T>C | HNPCC | Yes | 22.9 |
| 19 | F/75 | Gastric, 74 & 79; Breast, 77 | A-colon | MD | 1 | MSI-H | −/+/+/− | MLH1 | MLH1 c.1758dupC | No history | Yes | N/A |
| 20 | F/41 | Endometrial, 41 | D-colon | PD | 3 | MSI-H | +/−/−/nd | MSH2 | MSH2 c.1888G>T | Suspicious | Yes | 17.2 |
| 21 | M/23 | - | A-colon | MD | 3 | MSI-H | nd | Not done | MLH1 c.1758dupC | HNPCC | No | > 50 |
| 22 | M/35 | - | S-colon | PD | 3 | MSI-H | +/−/−/+ | MSH2 | MSH2 c.1129C>T | HNPCC | Yes | 47.2 |
| 23 | M/58 | - | Multiple | PD | 3 | MSI-H | −/+/+/− | MLH1 | MLH1 c.1758dupC | No history | Yes | N/A |
| 24 | M/23 | - | D-colon | MD | 4 | MSI-H | nd | Not done | MSH2 c.2653C>T | Suspicious | No | 35.0 |
| 25 | M/20 | - | A-colon | WD | 3 | MSI-H | −/+/+/− | MLH1 | MLH1 c.1721T>C | Suspicious | Yes | > 50 |
A-colon, ascending colon; D-colon, descending colon; HNPCC, hereditary nonpolyposis colorectal cancer; IHC, immunohistochemistry; MD, moderately differentiated; MMR, mismatch repair; MSI, microsatellite instability; MSI-H, microsatellite instability high; MSS, microsatellite stable; N/A, not available; nd, not done; PD, poorly differentiated; PREMM, prediction model for gene mutations; S-colon, sigmoid colon; T-colon, transverse colon; WD, well differentiated.
a) PREMM5 model prediction score was adapted from the Dana Farber Cancer Institute (http://premm.dfci.harvard.edu/).




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download