Abstract
Purpose
Appropriate preclinical mouse models are needed to evaluate the response to immunotherapeutic agents. Immunocompetent mouse models have rarely been reported for gastric cancer. Thus, we investigated immunophenotypes and responses to immune checkpoint inhibitor (ICI) in immunocompetent mouse models using various murine gastric cancer cell lines.
Materials and Methods
We constructed subcutaneous syngeneic tumors with murine gastric cancer cell lines, YTN3 and YTN16, in C57BL/6J mice. Mice were intraperitoneally treated with IgG isotype control or an anti–programmed death-ligand 1 (PD-L1) neutralizing antibody. We used immunohistochemistry to evaluate the tumor-infiltrating immune cells of formalin-fixed paraffin-embedded mouse tumor tissues. We compared the protein and RNA expression between YTN3 and YTN16 cell lines using a mouse cytokine array and RNA sequencing.
Results
The mouse tumors revealed distinct histological and molecular characteristics. YTN16 cells showed upregulation of genes and proteins related to immunosuppression, such as Ccl2 (CCL2) and Csf1 (M-CSF). Macrophages and exhausted T cells were more enriched in YTN16 tumors than in YTN3 tumors. Several YTN3 tumors were completely regressed by the PD-L1 inhibitor, whereas YTN16 tumors were unaffected. Although treatment with a PD-L1 inhibitor increased infiltration of T cells in both the tumors, the proportion of exhausted immune cells did not decrease in the non-responder group.
Gastric cancer is the fourth leading cause of cancer-related deaths worldwide due to the dismal prognosis of patients with metastatic gastric cancer [1]. Several targeted agents, as well as combined chemotherapy, have been applied to improve the outcomes but to no avail. Although immune checkpoint inhibitors (ICIs) were approved for gastric cancer several years ago, they only exhibited limited benefits for a significant proportion of patients [2], and the resistance mechanisms related to ICIs in gastric cancer remain unclear.
A better understanding of ICI resistance can suggest new targets for improving the prognosis for gastric cancer patients. The tumor microenvironment significantly affects the-rapeutic responses [3–5]. To investigate the exact mechanism related to ICI resistance and develop clinically efficacious therapeutics, we need experimental animal models. The widely used xenograft models do not reliably reflect the interaction between tumor cells and the host microenvironment. Syngeneic tumors in immunocompetent mice could be a good tool to evaluate the effectiveness of ICI in preclinical models [6]. Therefore, syngeneic murine models showing various responses to ICIs are essential for understanding the mechanisms of resistance to ICIs in gastric cancers. However, there are only a few reported syngeneic gastric cancer models [7,8].
Yamamoto et al. [9] developed murine gastric cancer cell lines from an N-methyl-N-nitrosourea (MNU)–induced gastric cancer mouse model. They reported that four cell lines from individual single-cell clones showed different tumorigenic potential and gene expression profiles. In the present study, we examined the histological and molecular features of two murine gastric cancer cell lines, in vivo and in vitro. In addition, we observed a distinct response to programmed death-ligand 1 (PD-L1) inhibitors in the two syngeneic gastric cancer models.
The aim of this study is to establish appropriate syngeneic mouse models to evaluate the effects of immunotherapy in gastric cancer and to identify the histological and molecular features of gastric cancers related to response to ICIs.
We cultured the murine gastric cancer cells, YTN3 and YTN16, in Dulbecco’s modified Eagle’s medium (DMEM)/high glucose (Cytiva, Marlborough, MA) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT), 1% penicillin-streptomycin (Gibco, Thermo Fisher Scientific, Waltham, MA), and 0.1% MITO+ serum extender (Corning, Corning, NY) on 0.5 mg/mL type I collagen (Corning) coated cell culture dish [9]. Cells were incubated at 37°C in a humidified atmosphere containing 5% CO2.
Five-week-old C57BL/6J mice (DBL, Eumseong, Korea) were subcutaneously injected with 1×106 YTN3 (n=10) or YTN16 (n=5) cells suspended in 100 μL phosphate buffered saline with 50% Matrigel Basement Membrane Matrix Growth Factor Reduced (Corning). The volume of the tumor was measured three times per week and calculated using the formula (length×width2)/2. We collected and weighed the tumor tissues and fixed them using 10% neutral buffered formalin (NBF), and embedded them in paraffin for further analysis.
For the treatment with PD-L1 antibody, the mice were injected with 1×106 YTN3 (n=15) or YTN16 (n=16) cells as described earlier. When the tumor volume reached 100 mm3, InVivoPlus anti-mouse PD-L1 (B7-H1) (BioXcell, Lebanon, PA) or InVivoMab rat IgG2b isotype control (BioXcell) was intraperitoneally administered once a week for three weeks (12.5 mg/kg). The volume of the tumor was measured three times per week. We collected and weighed the tumor tissues and fixed them using 10% NBF, and embedded them in paraffin for further analysis.
We cut the formalin-fixed paraffin-embedded tissues into 4 μm-thick sections and performed antigen retrieval using Tris-EDTA (pH 9.0)+10% glycerol solution. The slides were blocked in 20% Aqua Block Buffer solution (Abcam, Cambridge, UK) and incubated with the primary antibody overnight at 4°C. The slides were then incubated with the secondary antibody at room temperature for 30 minutes, followed by detection using the DAB Substrate Kit (Abcam). The antibodies used for immunohistochemistry were as follows: anti–α-smooth muscle actin (1:500, #14-9760-82, eBioscience, Thermo Fisher Scientific), anti-CD4 (1:2000, #ab183685, Abcam), anti-CD8α (1:4000, #98941, Cell Signaling Technology, Danvers, MA), anti-EOMES (1:500, #ab183991, Abcam), anti-F4/ 80 (1:500, #70076, Cell Signaling Technology), anti–PD-L1 (1:200, #64988, Cell Signaling Technology), and anti-rabbit (1:100, #Abc-5003, AbClon, Seoul, Korea). All antibodies were diluted in an Antibody Diluent for IHC (BD Biosciences, San Jose, CA).
All the stained slides were scanned using a slide scanner (Axioscan. Z1, Carl Zeiss Microscopy GmbH, Jena, Germany) at the Three-Dimensional Immune System Image Core Facility. Three regions of interest images for each tissue were acquired from the most intensively stained area, and the immunohistochemically stained positive cells were manually counted. This procedure was performed by two independent observers. The results are expressed as the mean number of positive cells per area (mm2). The percentage of exhausted immune cells is represented as a percentage of EOMES+ cells to CD8α+ or CD4+ cells.
Cells (5×105) were seeded in a collagen-coated 100-mm culture dish and cultured in 10 mL of serum-free DMEM/high glucose for 48 hours. The cell culture medium was collected into a 15 mL conical tube and centrifuged at 2,000 rpm, 4°C for 10 minutes. The supernatant was transferred to a new 15 mL conical tube and stored at −20°C. Cytokine levels in the conditioned medium were evaluated using the Proteome Profiler Mouse Cytokine Array Kit, Panel A (R&D Systems, Minneapolis, MN) following the manufacturer’s instructions. The pixel density of each spot was measured using ImageJ software.
Total RNA was extracted using an RNeasy Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s ins-tructions. Total RNA sequencing was performed using the NovaSeq platform (Illumina, San Diego, CA). Gene ontology (GO) analysis of differentially expressed genes (DEGs) was performed using the DAVID database [10].
Genomic DNA was extracted using a G-spin Total DNA extraction kit (iNtRON, Seongnam, Korea) following the manufacturer’s instructions. Whole-exome sequencing was performed using the Hiseq 2000, 2500, and 4000 (Illumina). Mm10 sequences were used as mapping references. Mutation and single nucleotide polymorphisms (SNPs) were analyzed using R package maftools.
Statistical analysis was performed using the R 4.1.1 software program and GraphPad Prism 9. Data are presented as mean±standard error. Two-way repeated-measures ANOVA was conducted to compare tumor volumes between the two groups. The means of the two groups were compared using a two-sample t test (parametric) or Wilcoxon rank-sum exact test (non-parametric). Statistical significance was set at p < 0.05.
First, to analyze the tumor microenvironment of the murine gastric cancer models, we subcutaneously injected murine gastric cancer cell lines into immunocompetent mice (Fig. 1A and B). On the 37th to 42nd day after cell injection, we harvested tumors and performed the hematoxylin and eosin stain. The pathologist (Lee SH) reviewed the slides and described that the cancer cells of YTN3 tumors formed the intestinal glands, which resembled the intestinal type and moderately differentiated gastric cancer (Fig. 1C). Meanwhile, YTN16 tumors, where cancer cells are scattered in the stroma, are similar to poorly differentiated and diffuse-type gastric cancer. We conducted immunohistochemical staining for several markers of immune cells and fibroblasts in the harvested tumor tissues (Fig. 1C–E). Tumor-infiltrating macrophages (F4/80+) and exhausted T cells (EOMES+) were significantly increased in YTN16 tumors, although the total number of CD8+ T cells (CD8α+) was not (Fig. 1D and E). Both tumors were positive for PD-L1 staining. These results indicate that YTN3 and YTN16 tumors have histologically distinct features and YTN16 tumors have a more immunosuppressive tumor microenvironment than YTN3 tumors.
Next, we investigated the molecular characteristics of these cancer cell lines that led to differences in the tumor microenvironment (Fig. 2A, S1 Fig.). In whole-exome sequencing data, YTN16 had a barely decreased number of mutations and SNPs compared to YTN3 (S1 Fig.). Transcriptome analysis showed that 1,503 protein-coding genes were upregulated in YTN16 cells, and 326 protein-coding genes were upregulated in YTN3 cells among the 2584 DEGs (|fold-change| ≥ 2, p < 0.05) (S2 Table). GO biological process terms indicated that terms related to epithelial to mesenchymal transition (EMT) were enriched in YTN16 cells, while the term cell differentiation was enriched in YTN3 cells (Fig. 2B). This result was consistent with the finding that YTN3 tumor showed moderately differentiated histological features. Additionally, terms associated with immunosuppression, such as the negative regulation of macrophage cytokine production and natural killer cell degranulation, were enriched in YTN16 cells. YTN16 cells showed increased positive regulation of interleukin-10 (IL-10) secretion, while YTN3 cells showed increased positive regulation of interferon-gamma (IFNγ) production. IL-10 is a key anti-inflammatory mediator that downregulates class II MHC expression and proinflammatory cytokine secretion [11]. IFNγ can stimulate hundreds of genes, including inflammatory molecules, apoptosis regulators, cell cycle regulators, and transcriptional activators [12].
We hypothesized that secreted proteins can modulate the tumor microenvironment. Therefore, we performed a cytokine array with conditioned media from each murine gastric cancer cell line. Eleven cytokines were differentially expressed between YTN3 and YTN16 cells (Fig. 2C). In the RNA sequencing data, the expression of the following genes encoding targeted proteins in the cytokine array was significantly changed (p < 0.05): Il1rn (IL-1ra), Ccl2 (CCL2), Csf1 (M-CSF), and Cxcl1 (CXCL1) (Fig. 2D). IL-1ra (interleukin-1 receptor antagonist), a natural inhibitor of IL-1α and IL-1β [13]; CCL2 (C-C motif chemokine ligand 2), also known as monocyte chemoattractant protein 1 (MCP1); and M-CSF (macrophage colony-stimulating factor) are highly expressed in YTN16 cells. CXCL1 (C-X-C motif chemokine ligand 1) was upregulated in YTN3 cells. Taken together, these mole-cular characteristics of cancer cell lines may be responsible for the distinct immunophenotypes in the tumor microenvironment.
To investigate the response to PD-L1 inhibitor in syngeneic mouse tumors, YTN3 and YTN16 cancer cells were transplanted subcutaneously into the right flank of mice. After the tumor volume reached 100 mm3, anti–PD-L1 antibody and IgG isotype control (12.5 mg/kg) were intraperitoneally administered (Fig. 3A–C). The PD-L1 inhibitor showed an antitumor effect only in YTN3 tumors. Moreover, four YTN3 tumors completely regressed (S3 Fig.). In contrast, the volume and weight of YTN16 tumors did not significantly differ with treatment (Fig. 3C). These results revealed that tumors constructed using two different murine gastric cancer cells with distinct molecular characteristics showed diverse res-ponses to PD-L1 inhibitor.
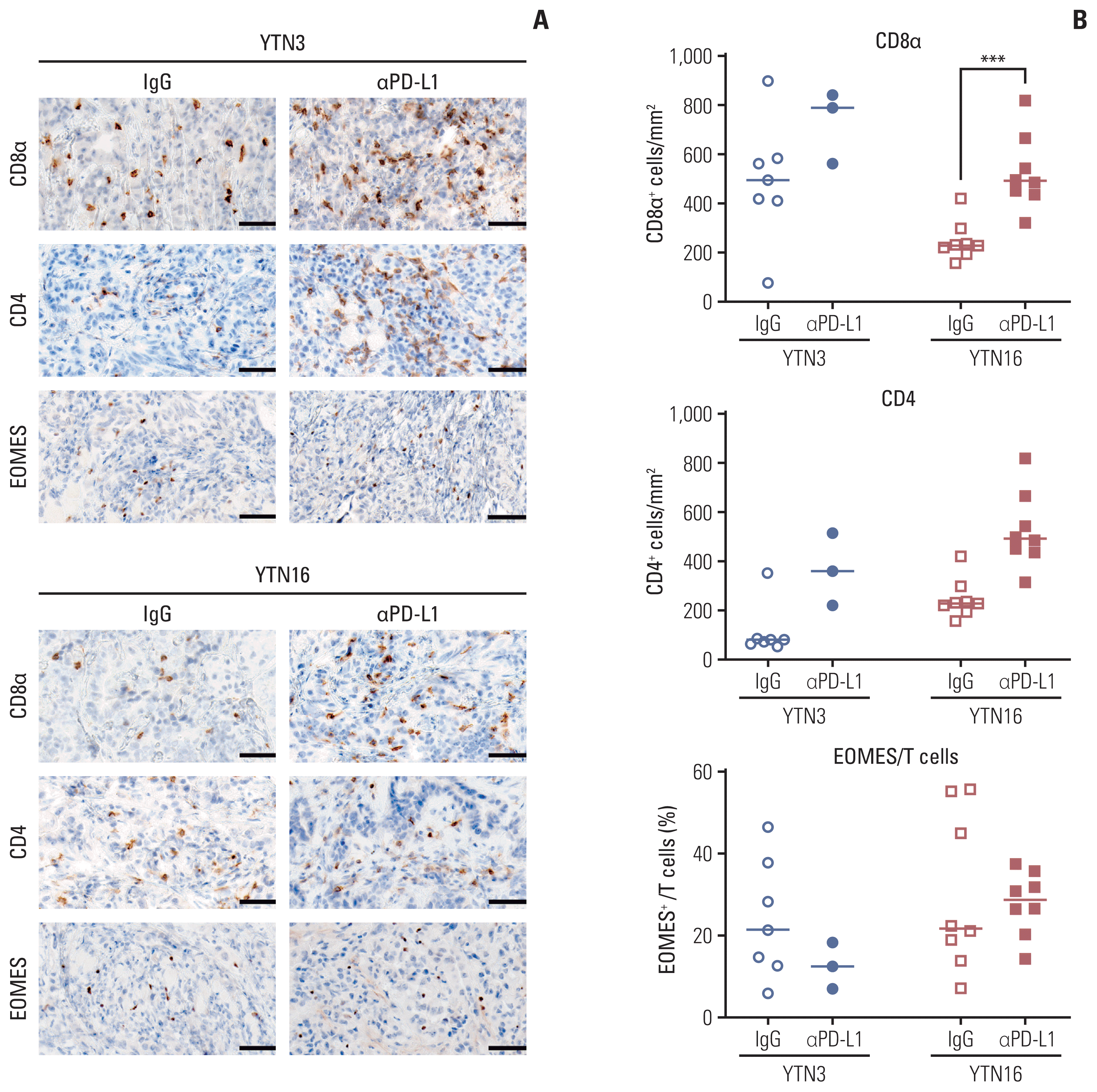
Finally, we confirmed the histological changes in the tumor microenvironment after PD-L1 inhibitor treatment using immunohistochemical staining (Fig. 4A and B). As expected, the number of tumor-infiltrating CD8+ T cells increased after treatment in both YTN3 and YTN16 tumors. Meanwhile, the number of CD4+ T cells was increased only in YTN3 tumors. The proportion of exhausted T cells to total T cells was lower in YTN3 tumors than in YTN16 tumors, but the difference was not significant. These results indicate that PD-L1 inhibitor treatment led to a reduced ratio of exhausted T cells and increased T cells in the responder group. As a result of the reprogrammed tumor microenvironment, tumor growth was effectively inhibited.
In this study, we established two syngeneic mouse models with different sensitivities to ICI. Notably, the two mouse models reproduced the different histological and molecular characteristics of human gastric cancers, associated with the response to PD-L1 inhibitor. YTN3 syngeneic tumors—that were significantly affected by PD-L1 inhibitor—demonstrated histological characteristics of intestinal-type gastric cancers, while YTN16 tumors exhibiting resistance to PD-L1 inhibitor showed the histologic feature of the diffuse-type tumors. Immunologic profiling of syngeneic tumors revealed that the proportion of immune cells expressing exhausted markers in YTN16 was higher than that in YTN3, and macrophages accumulated more in tumors resistant to ICI. Molecular analysis of two murine gastric cancer cell lines showed that YTN16 cells upregulated several gene sets correlated with the gene expression of diffuse-type gastric cancers and secreted several proteins related to immune evasion. Taken together, these results reveal the distinct characteristics of human gastric cancers related to the clinical response to ICIs.
In 2018, the Japanese group established a novel murine gastric cancer cell line, YTN, via chemical ingestion in immunocompetent mice [9]. Chemically induced primary gastric tumor tissues were primary cultured and the cells were subcutaneously injected into C57BL/6 mouse. YTN3 and YTN16 were established by different clones derived from the secondary subcutaneous tumor. There have been differences in tumorigenicity and molecular features between these two cell lines, but their immune profile and response to immune therapy have not been evaluated. Except for the YTN cell lines, other murine gastric cancer cell lines such as the S series and M12 developed by other researchers [7,8] have rarely been evaluated for immunotherapy response [14]. The present study revealed different molecular and immunologic profiles and histologic features of syngeneic tumors according to their response to ICIs.
Our study highlighted that molecular profiles of murine gastric cancer cells could be a cause of different histological features and immunological phenotypes in syngeneic tumors established with two different cell lines. Presumably, these molecular and histological features might lead to different responses to anti-PD-L1 treatment. The Cancer Genome Atlas data suggests that the four major molecular subtypes of gastric cancers have different histological characteristics [15]. Of the four subtypes, EBV or MSI gastric cancers prevalently covers intestinal subtypes and includes high levels of intratumoral CD8+ T cells [16]. Moreover, these subtypes have been suggested as gastric cancers that show a favorable response to anti–PD-L1 treatment [17]. Other high-throughput data for gastric cancers also showed that EMT subtypes showing poor clinical outcomes included a higher proportion of diffuse histologic tumors [18] that overlapped with non-EBV and non-MSI gastric cancers showing a poor response to anti–PD-L1 treatment. However, the mechanisms underlying the resistance of different histological and molecular subtypes to anti–PD-L1 treatment have not been elucidated. In the present study, YTN16, which showed a poor response to anti–PD-L1, showed upregulation of some gene sets related to mesenchymal transition of cancer cells and the histological features of diffuse-type cancer cells scattered into tumor stroma and not forming intestinal glands. In addition, in the whole-exome sequencing for those cell lines, YTN16 showed slightly a smaller number of mutations and SNPs compared to YTN3. The Cancer Genome Atlas data set revealed that low mutation burden was the main characteristic of genomically stable subset which enriched diffuse-type gastric cancer [15]. These results imply that YTN16 syngeneic tumors replicate the diffuse-type, including non-EBV and non-MSI subtypes. Our models could be useful to clarify the resistance mechanism according to the molecular and histological subtypes of gastric cancers in the future.
In the present study, an integrated analysis of secretome and transcriptome data for murine gastric cancer cell lines showed that YTN16 could secrete several factors, such as CCL2 and M-CSF. These secreted proteins might contribute to the suppressive immune microenvironment of YTN16 syngeneic tumors relative to that of YTN3 tumors, resulting in a poor response to ICIs in YTN16 syngeneic tumors. CCL2 is considered a crucial mediator between cancer cells and the tumor microenvironment. Several experimental studies have suggested that CCL2-induced enrichment of myeloid cells can promote tumor progression via the immune regulatory function of the host immune cells [19,20]. M-CSF can also enhance the recruitment, extravasation, and proliferation of monocytic precursor cells in primary tumors and consequently lead to their maturation into macrophages [21]. Our study showed that YTN16 tumors included a greater number of macrophages and exhausted T cells compared with YTN3 tumors, and CCL2 or M-CSF contribute to their immunologic phenotype. Macrophage-initiating immune evasion was explored as one of the reasons for poor response to ICIs in lung squamous cell carcinomas [22]. Our previous single-cell transcriptome data for diffuse-type gastric cancers suggested that an increased CCL2 gradient within the tumor microenvironment of the deep infiltrating region causes enrichment of tumor-associated macrophages and a suppressive immune environment [23]. Therefore, the present syngeneic mouse model might be useful for exploring macrophage-associated immune evasion and investigating the effect of immunotherapeutic agents targeting this mechanism.
Immunotherapies targeting inhibitory receptors such as cytotoxic T lymphocyte associated antigen 4 and programmed cell death-1 (PD-1) can successfully enhance T-cell function and achieve clinical effects in several solid tumors [24]. Clinical observations of patients treated with ICIs showed an influx of cytotoxic CD8+ T cells into the tumor microenvironment during treatment [25,26]. In the present study, anti–PD-L1 treatment provoked a tendency of more cytotoxic CD8+ T cell infiltration in responsive syngeneic tumors, but statistical significance was not proved due to the limited number of remaining samples after treatment. Intriguingly, even in the non-response group, anti–PD-L1 blockade significantly promoted the enrichment of CD8+ cytotoxic T cells. These results for our syngeneic tumors did not fit with previous clinical studies on melanoma, in which adaptive immune signatures in early on-time biopsied samples could predict the response to immune checkpoint blockade [27]. However, this previous study did not consider the diversity of T cell subpopulations and dynamic reprogramming during immunotherapy. Several previous studies have suggested that subpopulations of CD8+ T cells are related to alternative immune checkpoints that show a proliferative burst during anti–PD-L1 administration [28,29]. They demonstrated that the expansion of exhausted immune cell subpopulations can promote resistance to ICIs. In the present study, while the number of cytotoxic T cells increased during anti–PD-L1 treatment even in non-responsive YTN16 syngeneic tumors, the proportion of EOMES+ cells representing exhausted immune cells was not reduced. A previous study reported that EOMES expression in cytotoxic T cells could restrain CD8+ T cell antitumor function [30]. Taken together, we assume that the poor response of YTN16 syngeneic tumors to anti–PD-L1 treatment is caused by alternative immune checkpoints.
There are several limitations of the present study. First, we treated the mice with only PD-L1 inhibitor. Although PD-L1 monoclonal antibody, Avelumab, has tried to improve the survival of gastric cancer patients with metastasis in a third-line setting, the result was negative [31]. Meanwhile, PD-1 inhibitors like Nivolumab and Pembrolizumab showed the positive results for metastatic gastric cancer patients. To unravel the different responses of various ICIs in gastric cancer, further experiments using other ICIs would be needed. Second, we established subcutaneous models, but they could not precisely reflect natural course of gastric cancer like the metastasis into the peritoneum and the liver. The orthotopic models for gastric cancer were rarely reported because it was hard to inject the cells into the gastric wall without the leakage into the gastric lumen or the peritoneum [32,33]. Further experiments would be needed to unravel the different res-ponses to various ICIs in gastric cancer. Finally, the exact resistance mechanisms and ways to improve the efficacy of ICIs in non-responsive gastric cancer can be investigated using our gastric cancer syngeneic tumor model in the future.
In conclusion, we confirmed that the molecular features of cancer cells can lead to distinct responses to ICIs. A syngeneic gastric cancer murine model with resistance to PD-L1 inhibitor demonstrated distinct immune profiles, such as the enrichment of macrophages and exhausted immune cells. Our in vivo models can be useful for elucidating the mechanisms of resistance to ICIs to improve their clinical efficacy.
Electronic Supplementary Material
Supplementary materials are available at Cancer Research and Treatment website (https://www.e-crt.org).
Notes
Ethical Statement
Animal care and handling procedures were performed in accordance with the Ajou University School of Medicine Institutional Animal Care and Use Committee guidelines. All animal experiments were approved by the Animal Research Committee of the institution (2020-0043).
Acknowledgments
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education (2020R1A6A3A13071252 to Lee D; and 2020R1A6A1A03043539 to Hur H) and the Korean government, the Ministry of Science and ICT (MSIT) (2020R1A2C1006273 to Hur H).
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021; 71:209–49.
2. Kang YK, Boku N, Satoh T, Ryu MH, Chao Y, Kato K, et al. Nivolumab in patients with advanced gastric or gastro-oesophageal junction cancer refractory to, or intolerant of, at least two previous chemotherapy regimens (ONO-4538-12, ATTRACTION-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017; 390:2461–71.
3. Wu T, Dai Y. Tumor microenvironment and therapeutic response. Cancer Lett. 2017; 387:61–8.
4. Hirata E, Sahai E. Tumor microenvironment and differential responses to therapy. Cold Spring Harb Perspect Med. 2017; 7:a026781.
5. Roma-Rodrigues C, Raposo LR, Cabral R, Paradinha F, Baptista PV, Fernandes AR. Tumor microenvironment modulation via gold nanoparticles targeting malicious exosomes: implications for cancer diagnostics and therapy. Int J Mol Sci. 2017; 18:162.
6. Chulpanova DS, Kitaeva KV, Rutland CS, Rizvanov AA, Solovyeva VV. Mouse tumor models for advanced cancer immunotherapy. Int J Mol Sci. 2020; 21:4118.
7. Hsu HP, Wang CY, Hsieh PY, Fang JH, Chen YL. Knockdown of serine/threonine-protein kinase 24 promotes tumorigenesis and myeloid-derived suppressor cell expansion in an orthotopic immunocompetent gastric cancer animal model. J Cancer. 2020; 11:213–28.
8. Park JW, Um H, Yang H, Ko W, Kim DY, Kim HK. Proteogenomic analysis of NCC-S1M, a gastric cancer stem cell-like cell line that responds to anti-PD-1. Biochem Biophys Res Commun. 2017; 484:631–5.
9. Yamamoto M, Nomura S, Hosoi A, Nagaoka K, Iino T, Yasuda T, et al. Established gastric cancer cell lines transplantable into C57BL/6 mice show fibroblast growth factor receptor 4 promotion of tumor growth. Cancer Sci. 2018; 109:1480–92.
10. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009; 4:44–57.
11. Saraiva M, Vieira P, O’Garra A. Biology and therapeutic potential of interleukin-10. J Exp Med. 2020; 217:e20190418.
12. Castro F, Cardoso AP, Goncalves RM, Serre K, Oliveira MJ. Interferon-gamma at the crossroads of tumor immune surveillance or evasion. Front Immunol. 2018; 9:847.
13. Perrier S, Darakhshan F, Hajduch E. IL-1 receptor antagonist in metabolic diseases: Dr Jekyll or Mr Hyde? FEBS Lett. 2006; 580:6289–94.
14. Nagaoka K, Shirai M, Taniguchi K, Hosoi A, Sun C, Kobayashi Y, et al. Deep immunophenotyping at the single-cell level identifies a combination of anti-IL-17 and checkpoint blockade as an effective treatment in a preclinical model of data-guided personalized immunotherapy. J Immunother Cancer. 2020; 8:e001358.
15. Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014; 513:202–9.
16. De Rosa S, Sahnane N, Tibiletti MG, Magnoli F, Vanoli A, Sessa F, et al. EBV(+) and MSI gastric cancers harbor high PD-L1/PD-1 expression and high CD8(+) intratumoral lymphocytes. Cancers (Basel). 2018; 10:102.
17. Rodriquenz MG, Roviello G, D’Angelo A, Lavacchi D, Roviello F, Polom K. MSI and EBV positive gastric cancer’s subgroups and their link with novel immunotherapy. J Clin Med. 2020; 9:1427.
18. Cristescu R, Lee J, Nebozhyn M, Kim KM, Ting JC, Wong SS, et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med. 2015; 21:449–56.
19. Kersten K, Coffelt SB, Hoogstraat M, Verstegen NJ, Vrijland K, Ciampricotti M, et al. Mammary tumor-derived CCL2 enhances pro-metastatic systemic inflammation through upregulation of IL1beta in tumor-associated macrophages. Oncoimmunology. 2017; 6:e1334744.
20. Lee GT, Kwon SJ, Kim J, Kwon YS, Lee N, Hong JH, et al. WNT5A induces castration-resistant prostate cancer via CCL2 and tumour-infiltrating macrophages. Br J Cancer. 2018; 118:670–8.
21. Van Overmeire E, Stijlemans B, Heymann F, Keirsse J, Morias Y, Elkrim Y, et al. M-CSF and GM-CSF receptor signaling differentially regulate monocyte maturation and macrophage polarization in the tumor microenvironment. Cancer Res. 2016; 76:35–42.
22. Peranzoni E, Lemoine J, Vimeux L, Feuillet V, Barrin S, Kantari-Mimoun C, et al. Macrophages impede CD8 T cells from reaching tumor cells and limit the efficacy of anti-PD-1 treatment. Proc Natl Acad Sci U S A. 2018; 115:E4041–50.
23. Jeong HY, Ham IH, Lee SH, Ryu D, Son SY, Han SU, et al. Spatially distinct reprogramming of the tumor microenvironment based on tumor invasion in diffuse-type gastric cancers. Clin Cancer Res. 2021; 27:6529–42.
24. Sharma P, Wagner K, Wolchok JD, Allison JP. Novel cancer immunotherapy agents with survival benefit: recent successes and next steps. Nat Rev Cancer. 2011; 11:805–12.
25. Huang RR, Jalil J, Economou JS, Chmielowski B, Koya RC, Mok S, et al. CTLA4 blockade induces frequent tumor infiltration by activated lymphocytes regardless of clinical responses in humans. Clin Cancer Res. 2011; 17:4101–9.
26. Ng Tang D, Shen Y, Sun J, Wen S, Wolchok JD, Yuan J, et al. Increased frequency of ICOS+ CD4 T cells as a pharmacodynamic biomarker for anti-CTLA-4 therapy. Cancer Immunol Res. 2013; 1:229–34.
27. Chen PL, Roh W, Reuben A, Cooper ZA, Spencer CN, Prieto PA, et al. Analysis of immune signatures in longitudinal tumor samples yields insight into biomarkers of response and mechanisms of resistance to immune checkpoint blockade. Cancer Discov. 2016; 6:827–37.
28. Im SJ, Hashimoto M, Gerner MY, Lee J, Kissick HT, Burger MC, et al. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature. 2016; 537:417–21.
29. Koyama S, Akbay EA, Li YY, Herter-Sprie GS, Buczkowski KA, Richards WG, et al. Adaptive resistance to therapeutic PD-1 blockade is associated with upregulation of alternative immune checkpoints. Nat Commun. 2016; 7:10501.
30. Weulersse M, Asrir A, Pichler AC, Lemaitre L, Braun M, Carrie N, et al. Eomes-dependent loss of the co-activating receptor CD226 restrains CD8(+) T cell anti-tumor functions and limits the efficacy of cancer immunotherapy. Immunity. 2020; 53:824–39.
31. Bang YJ, Ruiz EY, Van Cutsem E, Lee KW, Wyrwicz L, Schenker M, et al. Phase III, randomised trial of avelumab versus physician’s choice of chemotherapy as third-line treatment of patients with advanced gastric or gastro-oesophageal junction cancer: primary analysis of JAVELIN Gastric 300. Ann Oncol. 2018; 29:2052–60.
32. Kang W, Maher L, Michaud M, Bae SW, Kim S, Lee HS, et al. Development of a novel orthotopic gastric cancer mouse model. Biol Proced Online. 2021; 23:1.
33. Song S, Xu Y, Huo L, Zhao S, Wang R, Li Y, et al. Patient-derived cell lines and orthotopic mouse model of peritoneal carcinomatosis recapitulate molecular and phenotypic features of human gastric adenocarcinoma. J Exp Clin Cancer Res. 2021; 40:207.
Fig. 1
Histological features of syngeneic mouse tumors. (A) 1×106 of YTN3 (n=10) and YTN16 (n=5) cells were subcutaneously injected into C57BL/6J mice. (B) Tumor volume and weight of YTN3 (top) and YTN16 (bottom) tumors. Values are presented as mean±standard error of the mean (SEM). (C) Representative images of H&E staining and immunohistochemistry for α-smooth muscle actin (α-SMA) (×200, scale bars=50 μm). (D) Representative images for immunohistochemistry; CD8α for cytotoxic T cell, F4/80 for macrophage, EOMES for exhaustion marker, and programmed death-ligand 1 (PD-L1) for immune checkpoint (×200, scale bars=50 μm). (E) Immunohistochemistry staining. Positive cells were counted per mm2. **p < 0.01, ****p < 0.0001.
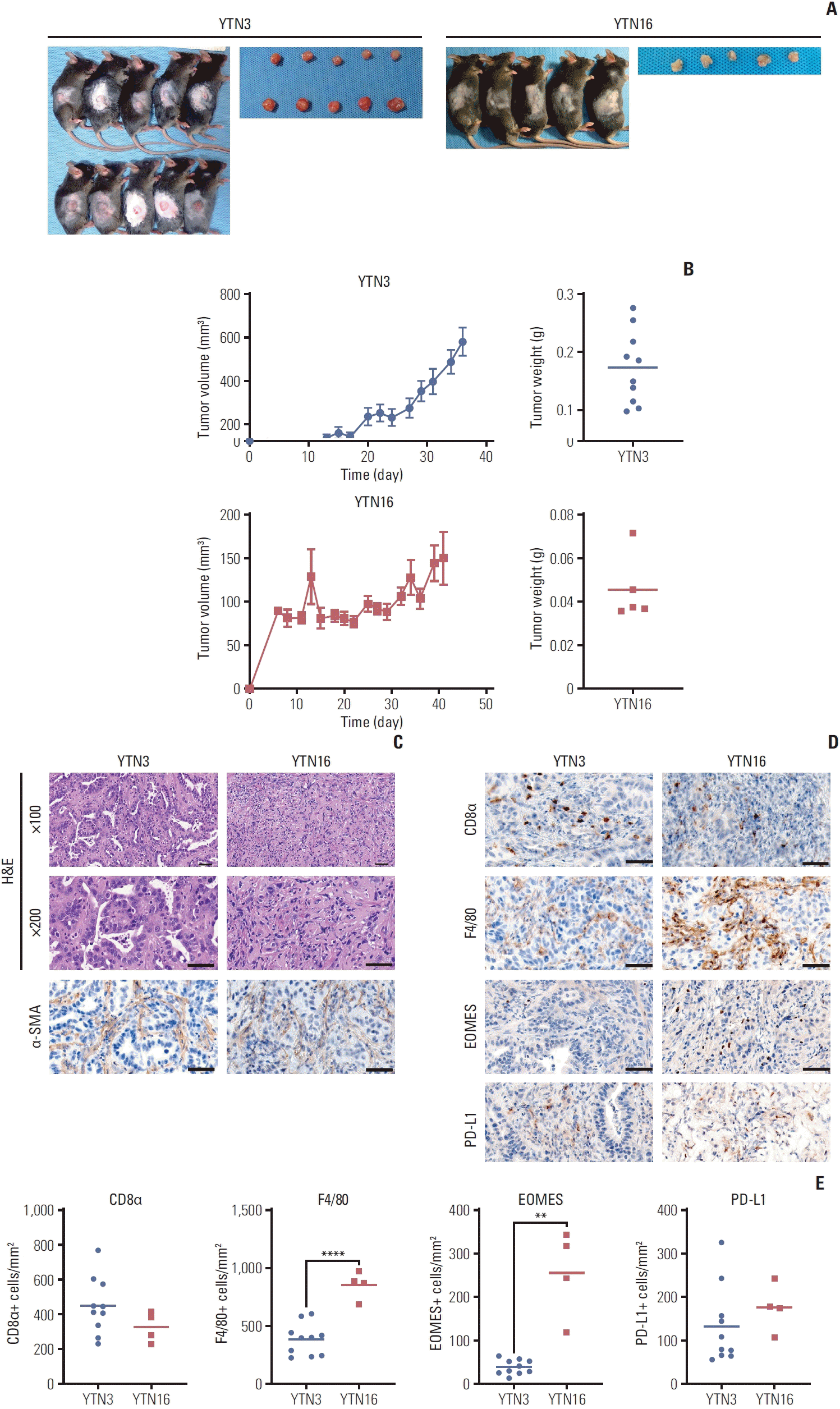
Fig. 2
Molecular characteristics of YTN3 and YTN16 murine gastric cancer cell lines. (A) Schematic figure for RNA-sequencing and mouse cytokine array workflow. (B) Gene ontology (GO) biological process analysis for differentially expressed genes (|fold-change| ≥ 2, p < 0.05) using DAVID database. *p < 0.05, **p < 0.01, ***p < 0.001. (C) Mouse cytokine array panels for conditioned media of YTN3 and YTN16 cells (left). Mean pixel density was quantified for each target protein (right). (D) The expression of genes coding cytokine array–targeted proteins from RNA sequencing. *p < 0.05, ***p < 0.001.
Fig. 3
Response to programmed death-ligand 1 (PD-L1) inhibitor in syngeneic mouse tumors. (A) The schematic figure for anti–PD-L1 antibody treatment in vivo. 1×106 of YTN3 (n=15) and YTN16 (n=16) cells were subcutaneously injected in C57BL/6J mice. IgG isotype control or anti–PD-L1 antibody (12.5 mg/kg) was administered once a week, intraperitoneally. (B) Subcutaneous tumors of YTN3 and YTN16 cell lines after anti–PD-L1 antibody treatment. (C) Tumor volume and weight of YTN3 (top) and YTN16 (bottom) tumors. Values are presented as mean±standard error of the mean (SEM). **p < 0.01; ns, not significant.
Fig. 4
Reprograming of tumor microenvironment after programmed death-ligand 1 (PD-L1) inhibitor treatment. (A) Representative images for immunohistochemistry; CD8α, CD4, and EOMES (×200, scale bars=50 μm). (B) Immunohistochemistry. Positive cells were counted per mm2. The percentage of exhausted immune cells is represented as a percentage of EOMES+ cells to CD8α+ or CD4+ cells. ***p < 0.001.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download