INTRODUCTION
Upon the outbreak of coronavirus disease 2019 (COVID-19) caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the World Health Organization (WHO) declared a pandemic on March 11, 2020. Many countries remain disrupted by this virus and still endure COVID-19
(1,
2,
3). Moreover, SARS-CoV-2 variants are emerging in various parts of the world, with some strains displaying even greater infectivity and transmissibility
(4,
5).
In the early days of the pandemic, the understanding of COVID-19 and its therapeutic management was limited; it was thought that experimental therapies and drug repurposing should be used for its alleviation. This naivety propelled intense efforts from researchers globally, which yielded significant progress, has led to a better understanding of COVID-19 and its management and resulted in the development of novel therapeutics and vaccines at an unprecedented speed
(6,
7,
8).
Currently, antiviral therapy that can be administered early after diagnosis and therapy to improve outcomes are provided to patients at risk of developing a severe form of the disease
(9). These treatments are divided into direct-acting antivirals (DAAs) and host-targeting agents (HTAs), where DAAs target a component of the virus and inhibit its replication
(10) and HTAs comprises many host factors involved in not only regulation of inflammatory responses but also viral replication, gene expression, assembly, and exit
(11,
12).
Various therapeutic options, including antiviral drugs (e.g., molnupiravir, paxlovid, remdesivir [RDV])
(13,
14), anti-SARS- CoV-2 monoclonal antibodies (e.g., bamlanivimab/etesevimab, casirivimab/imdevimab)
(15,
16), anti-inflammatory drugs (e.g., dexamethasone)
(17,
18), and immunomodulators (e.g., baricitinib and tocilizumab)
(19,
20), are available under the Food and Drug Administration (FDA)-issued Emergency Use Authorization (EUA) or are being evaluated for the management of COVID-19
(21,
22).
Drug screening is necessary to select and rapidly define potential treatments and could be a practical approach for testing and validating antiviral efficacy
(23). Although many studies on drug therapy for COVID-19 are available, the compatibility of each research method and system is unclear, limiting the consensus on COVID-19 treatment
(22). Therefore, selecting candidate therapeutics by accurately evaluating antiviral efficacy may facilitate the recommendation and/or implementation of drugs with effective anti-COVID-19 activity.
To this end, in the present study, we established
in vitro methods for plaque and tissue culture infectious dose 50 (TCID
50) assays and real-time quantitative reverse transcription (qRT)-PCR to evaluate the antiviral effects of a drug on COVID-19. These assays were performed using remdesivir (RDV), which has been previously reported to exhibit antiviral activity
(24,
25,
26), and were validated by testing plaque reduction or cytopathic effect (CPE) in cell culture against SARS-CoV-2 and the efficiency of viral gene replication
in vitro.
Go to :

MATERIALS AND METHODS
Cells and viruses
Vero E6 cells (KCLB no. 21587) were purchased from the Korean Cell Line Bank (Seoul, Republic of Korea) and maintained in Dulbecco’s modified Eagle’s medium (DMEM; Thermo Fisher Scientific, MA, USA) containing 10% fetal bovine serum (FBS; Thermo Fisher Scientific) and 1% penicillin/streptomycin (P/S; Thermo Fisher Scientific).
Table 1 shows SARS-CoV-2 and its variants of concern used in this study. Six SARS-CoV-2 variants, including SARS-CoV-2 (2019-nCoV, NCCP no. 43326), SARS-CoV-2 (B.1.1.7, NCCP no. 43381), SARS-CoV-2 (B.1.351, NCCP no. 43382), SARS-CoV-2 (P.1, NCCP no. 43388), SARS-CoV-2 (B.1.617.2, NCCP no. 43390), and SARS-CoV-2 (BA.2, NCCP no. 43412), which were provided by the National Culture Collection for Pathogens (NCCP; Osong, Korea), were prepared by propagation in Vero E6 cells. Cell culture procedures were performed at biosafety level 2 (BSL2) and moved to a BSL3 laboratory for viral infection and assays. All virus culture and assays were carried out in the BSL3 facility at the Konkuk University (Seoul, Korea).
Table 1.
SARS-CoV-2 variants of concern
|
WHO label
|
Clade
|
Lineage
|
Country first detected
|
NCCP no.
|
|
2019-nCoV
|
S
|
|
China
|
43326
|
|
Alpha
|
GRY
|
B.1.1.7
|
United Kingdom
|
43381
|
|
Beta
|
GH
|
B.1.351
|
South Africa
|
43382
|
|
Gamma
|
GR
|
P.1
|
Brazil
|
43388
|
|
Delta
|
GK
|
B.1.617.2
|
India
|
43390
|
|
Omicron
|
GRA
|
BA.2
|
Southern Africa
|
43412
|

Chemicals
RDVs (cat. HY-104077) were purchased from MedChemExpress (NJ, USA) and dissolved in dimethyl sulfoxide (DMSO; Sigma-Aldrich) to a final concentration of 10 mM.
Cytotoxicity assay
To determine the cytotoxicity of pine needle extracts, cell viability was measured using the water-soluble tetrazolium salt method with an EZ-Cytox kit (Daeil Lab Service, Korea) according to the manufacturer’s instructions. Vero E6 cells were seeded in a 96-well plate at 1 × 104 cells/well density. The cells were cultured for 1 day, treated with serial dilutions of the extracts, and incubated at 37°C for 2 days. EZ-Cytox solution was added to each well and incubated for 2 h, followed by a spectrophotometric measurement of the absorbance at 540 nm. The 50% cytotoxic concentration (CC50) was calculated using GraphPad Prism 9 software (GraphPad, CA, USA).
Plaque reduction assay
A plaque assay was performed to determine the infectious viral titers of SARS-CoV-2 and its variants. A confluent monolayer of Vero E6 cells was prepared and inoculated with 100 plaque-forming units (PFU) of viruses in a 6-well plate. The viral inoculum was removed, and the cells were treated with ten-fold serially diluted chemicals for 1 h. Control wells were treated with phosphate-buffered saline (PBS), after which the chemical mixture was removed, and the cells were overlaid with DMEM (HyClone, UT, USA) containing 1.5% agarose (Lonza, MA, USA). After 72 h of treatment, diluted RDV and agarose were mixed and overlaid on the cells (
Fig. 1). After 72 h at 37°C and 5% CO
2, cells were stained with crystal violet (Georgia Chemicals, GA, USA).
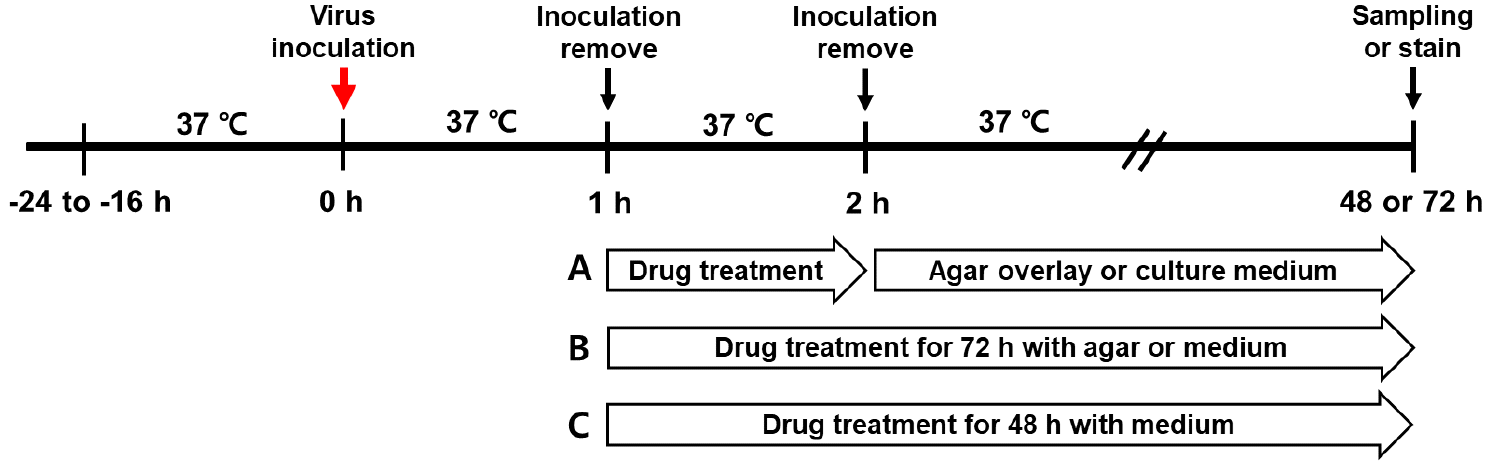 | Fig. 1Experimental mechanism of the SARS-CoV-2 infection and drug treatment. Vero E6 cells were inoculated with the SARS-CoV-2 variants for 1 h. After removing the viral inoculum, cells were incubated with remdesivir (RDV) at 37°C and 5% CO2. (A) For the plaque assay, cells were treated with RDV for 1 h, and the agar-overlay or the drug mixture was cultured with agarose overlay for 72 h to observe virus-induced plaques. (B) Cell treated with RDV for 1 or 72 h for TCID50 assays to observe CPE. (C) drug treatment for 48 h to detect viral RNA in the culture supernatants by qRT-PCR.
|
Tissue culture infectious dose (TCID50)
A day before infection, 1 × 10
4 cells/well were seeded in a 96-well plate and incubated overnight, after which the cells were inoculated with 50 TCID
50/well of viruses in a 96-well plate. The viral inoculum was removed, and the cells were treated with ten-fold serially diluted chemicals. After 1 h, the chemical mixture was removed, and the cells were incubated with 3% FBS culture media for 72 h at 37°C and 5% CO
2. After 72 h of treatment, the cells were cultured by mixing the diluted RDV and culture medium (
Fig. 1). The inoculum was removed, and the cells were stained for 30 min with 0.2% crystal violet solution to reveal CPEs. The crystal violet solution was removed, and the plates were washed with tap water and fully dried at room temperature. Fifty percent of endpoints were calculated using the Spearman-Karber calculation and expressed as TCID
50/ml
(27). Three independent experiments with six replicates were performed to determine viral titers by TCID
50.
Real-time quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
Vero E6 cells were inoculated with 50 PFU/well of the virus in a 12-well plate. After 1 h, the viral inoculum was removed and treated with 1 µM RDV. The cells were incubated in a 3% FBS culture medium for 48 h at 37°C and 5% CO2. The cell culture supernatant (100 µL) was harvested for viral RNA isolation using a NucleoSpi kit (Macherey-Nagel, PA, USA). Total RNA (1 µg) was reverse-transcripted into complementary DNA (cDNA) using SuperScript II reverse transcriptase (Invitrogen, CA, USA). qRT-PCR was performed using a TB Green Kit (TAKARA, Germany). The cycling parameters of the qPCR were as follows: 1 cycle at 94°C for 3 min, followed by 45 cycles at 94°C for 15 s and 60°C for 30 s in a real-time PCR instrument (Applied Biosystems™, 7500 Fast Real-Time PCR System; Thermo Fisher Scientific, Waltham, MA, USA). The SARS-CoV-2 nucleocapsid (N) region was amplified using forward (5’-AAATTTGGGGACCAGGAAC) and reverse (5’-TGGCAGCTGTG TAGGTCAAC) primers.
Statistical analysis
All experiments were carried out three times independently at two laboratories. All statistical analyses were performed using GraphPad Prism 9 (GraphPad). The significance of differences between treatment groups was analyzed using a two-way analysis of variance (ANOVA) or two-tailed Student’s t-test. P values < 0.05 were considered statistically significant.
Go to :

RESULTS
Plaque inhibitory effect of RDV using plaque assay
The virus titer was determined by plaque reduction and TCID50 assays, and the average number of viral stocks was calculated as 9.52×105 PFU/ml by plaque assays or 8.15×106 TCID50 /ml by TCID50 assay. Both assays exhibited no cytotoxicity in Vero E6 cells, and the CC50 of RDV, used as a therapeutic substance for COVID-19, was greater than 100 µM (data not shown).
To test the antiviral effect of RDV against SARS-CoV-2, the established plaque assay was used (
Fig. 2A). The antiviral activity of RDV against each of the six SARS-CoV-2 variants was confirmed through plaque reduction; a strong dose-dependent reduction in plaque levels was observed in the RDV-treated cells.
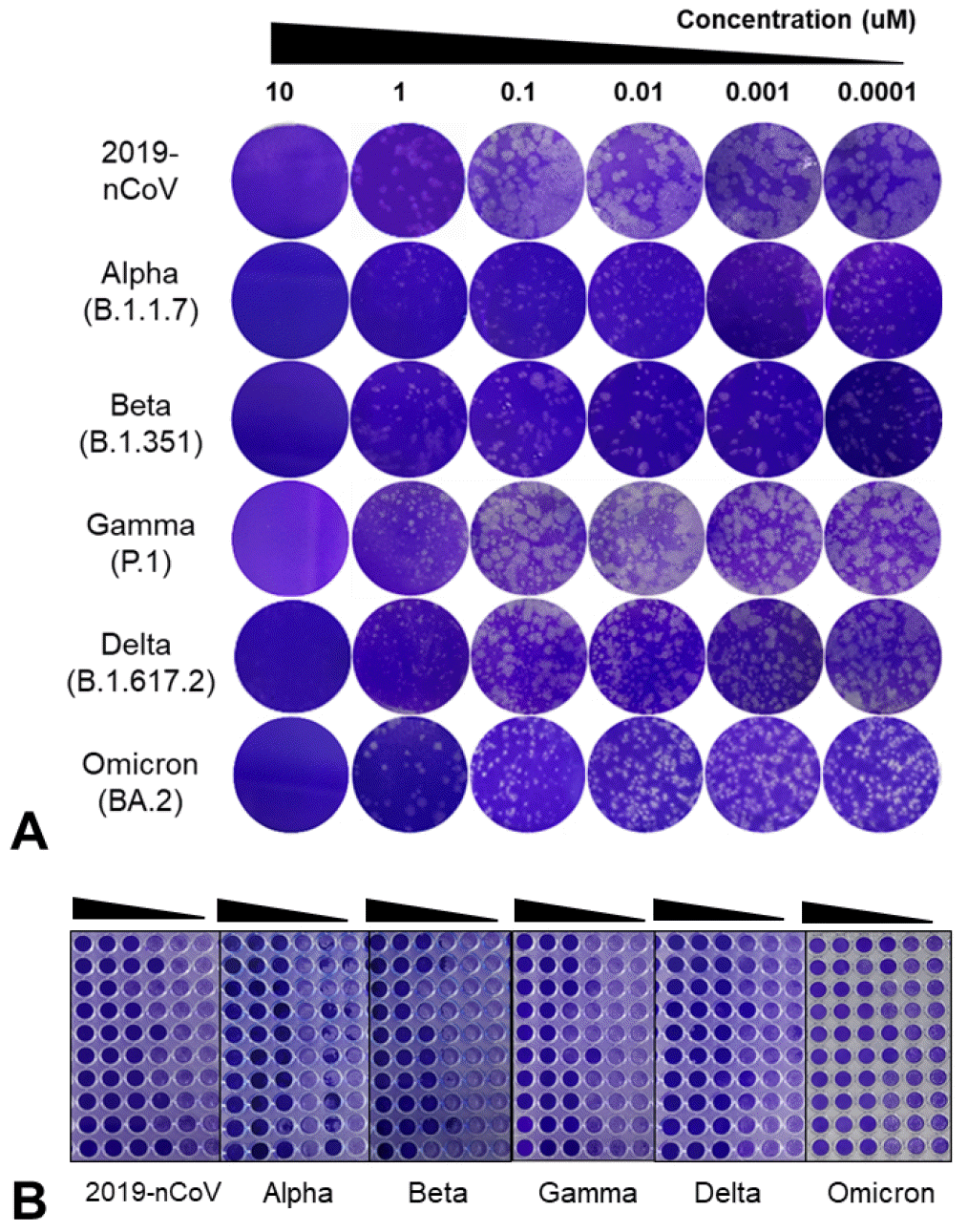 | Fig. 2Inhibitory effects of remdesivir (RDV) against SARS-CoV-2 variants. RDV was screened for its anti-SARS-CoV-2 efficacy using a plaque reduction assay (A), and TCID50 assay (B). Vero E6 cells were treated with RDV for 72 h in 10-fold serial dilutions (10, 1, 0.1, 0.01, 0.001, and 0.0001 µM). Statistical significance was determined using the multiple Student’s t-tests (P < 0.01).
|
First, 100 PFU/well of the virus was treated with RDV for 1 h after viral infection. At 10 µM RDV concentration, each variant was inhibited to a level of 50-100%, whereas an RDV concentration of ≤ 0.001 µM did not exert an inhibitory effect on all six variants (
Fig. 3A). The 50% inhibitory concentration (IC
50) was 2.17, 5.08, 5.82, 9.8, 9.8, and 9.1 uM for 2019-nCoV, SARS-CoV-2 B.1.17 (Alpha), SARS-CoV-2 B.1.351 (Beta), SARS-CoV-2 P.1 (Gamma), SARS-CoV-2 B.1.617.2 (Delta), and SARS-CoV-2 BA.2 (Omicron), respectively (
Table 2). Second, RDV treatment for 72 h after viral infection revealed that an RDV concentration of ≥ 1 µM exerted a 100% inhibitory effect on all six variants (
Fig. 3C). The IC
50 of 2019-nCoV, Alpha, Beta, Gamma, Delta, and Omicron was 0.22, 0.21, 0.28, 0.31, 0.32, and 0.35 uM, respectively (
Table 2).
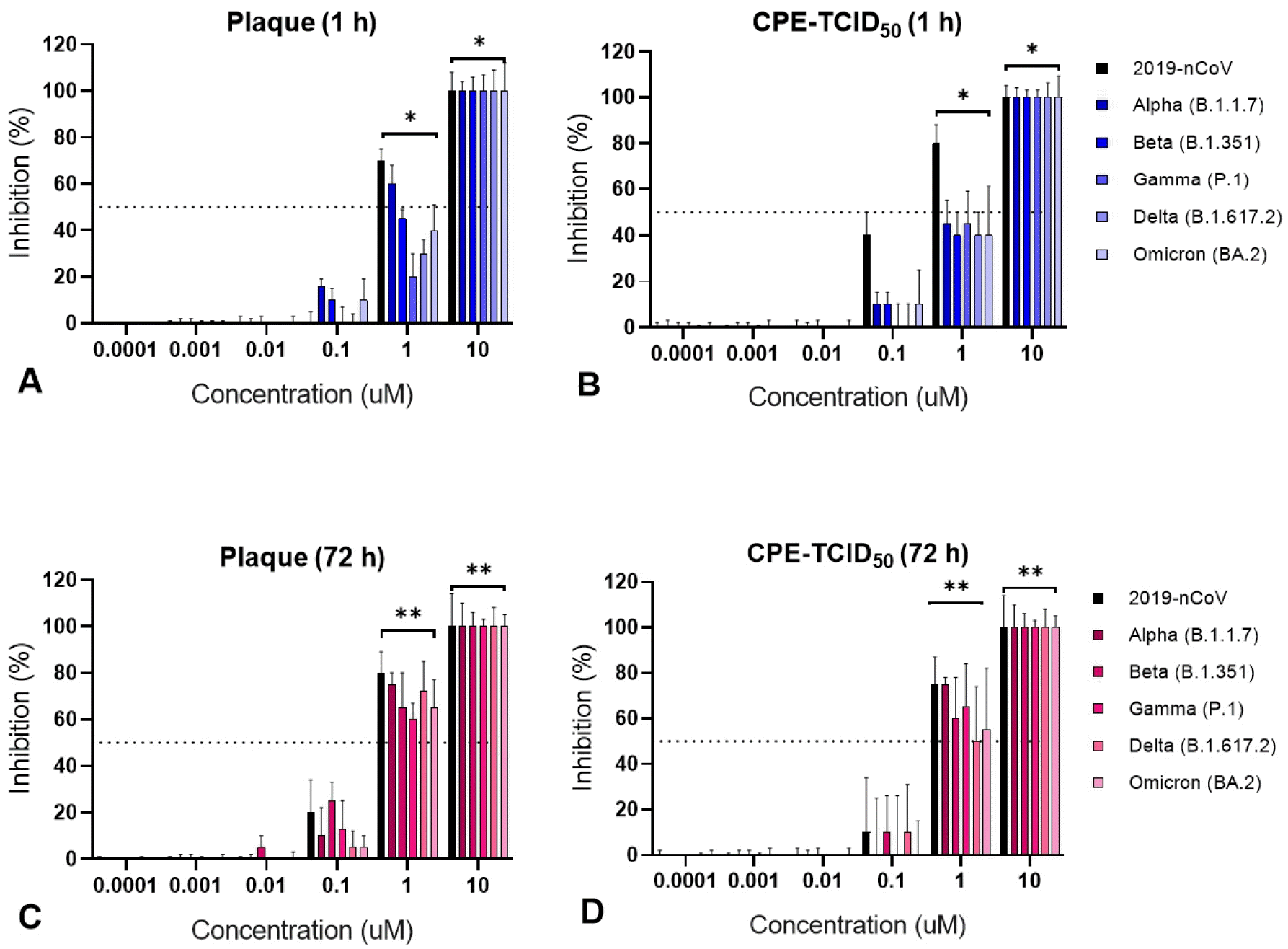 | Fig. 3Antiviral effect according to remdesivir (RDV) treatment time and concentration. RDV was screened for its anti-SARS- CoV-2 efficacy plaque reduction and TCID50 assays. Inhibition of SARS-CoV-2 variants was compared by treating cells with each concentration of RDV (0.0001, 0.001, 0.01, 0.1, 1, and 10 µM) for 1 h (A and B), or 72 h (C and D). Statistical analysis was performed using ANOVA and GraphPad Prism (*p < 0.05, **p<0.01).
|
Table 2.
Inhibitory effects of remdesivir against SARS-CoV-2 variants
|
|
IC50* (uM) by plaque
|
|
IC50 (uM) by TCID50
|
|
SARS-CoV-2
|
1 h treat
|
72 h treat
|
|
1 h treat
|
72 h treat
|
|
2019-nCoV
|
2.17
|
0.22
|
|
2.0
|
0.32
|
|
Alpha (B.1.1.7)
|
5.08
|
0.21
|
|
6.9
|
0.32
|
|
Beta (B.1.351)
|
5.82
|
0.28
|
|
7.4
|
0.50
|
|
Gamma (P.1)
|
9.8
|
0.31
|
|
9.2
|
0.40
|
|
Delta (B.1.617.2)
|
9.8
|
0.32
|
|
9.6
|
0.59
|
|
Omicron (BA.2)
|
9.1
|
0.35
|
|
9.8
|
0.51
|

Regardless of the viral strain, the median IC50 at treatment with RDV for 1 h and 72 h was 7.06 uM (± 3.21) and 0.28 uM (± 0.06), respectively. The plaque analysis data were slightly more broadly distributed around the median and had a wider 95% confidence interval (CI) than the TCID50 analysis data. The antiviral effect of RDV with reduced plaque levels was approximately 25-fold higher in the 72 h treatment than in the 1 h treatment (P < 0.001). In addition, RDV-induced plaque suppression was more effective against 2019-nCoV than other variants.
Inhibitory CPE of RDV using a TCID50 assay
To analyze the antiviral activity of RDV against SARS-CoV-2, the established TCID
50 assay was used. Cells in the 96-well plate were infected with 50 TCID
50/well of the viruses and treated with ten-fold serially diluted RDV (
Fig. 2B) for 1 or 72 h. A strong dose-dependent reduction in CPE levels was observed in the RDV-treated cells.
RDV treatment for 1 h after viral infection resulted in an IC
50 of 2.0, 6.9, 7.4, 9.2, 9.6, and 9.8 uM for 2019-nCoV, Alpha, Beta, Gamma, Delta, and Omicron, respectively. The 72 h RDV treatment resulted in an IC
50 of 0.32, 0.32, 0.5, 0.4, 0.59, and 0.51 uM for 2019-nCoV, Alpha, Beta, Gamma, Delta, and Omicron, respectively, similar to the plaque reduction test results (
Table 2).
The median IC50 at RDV treatment for 1 and 72 h was 8.25 uM (± 3.17) and 0.40 uM (± 0.25), respectively. The antiviral effect of RDV with reduced levels of CPE was approximately 20 times higher in the 72 h treatment than in the 1 h treatment (P < 0.001). The antiviral effect of RDV in Vero E6 cells was more effective against 2019-nCoV than the other variants.
The plaque and TCID50 assays revealed that RDV exerted the highest level of inhibition against 2019-nCoV in cells, and its antiviral effect was directly proportional to the treatment time and dose.
Replication inhibitory effect of RDV using qRT-PCR
To evaluate the antiviral efficacy of RDV against SARS-CoV-2, RDV was investigated using a cell-based gene copy number assay. Uninfected and infected cells were treated with RDV for 48 h. SARS-CoV-2 RNA levels were quantified by qRT-PCR targeting the N coding region.
A decrease in copy number was observed in qRT-PCR targeting the SARS-CoV-2 N gene (
Fig. 4). RDV inhibited viral RNA synthesis of each variant in Vero E6 cells. At 1 μM, RDV had a statistically significant antiviral effect (p < 0.001) on SARS-CoV-2 replication at 48 h of treatment (
Fig. 4A). Compared with the reduction level in the non-treated control group, the RDV-treated group showed an inhibition rate of approximately 90% for each virus (99% for 2019-nCoV) (
Fig. 4B).
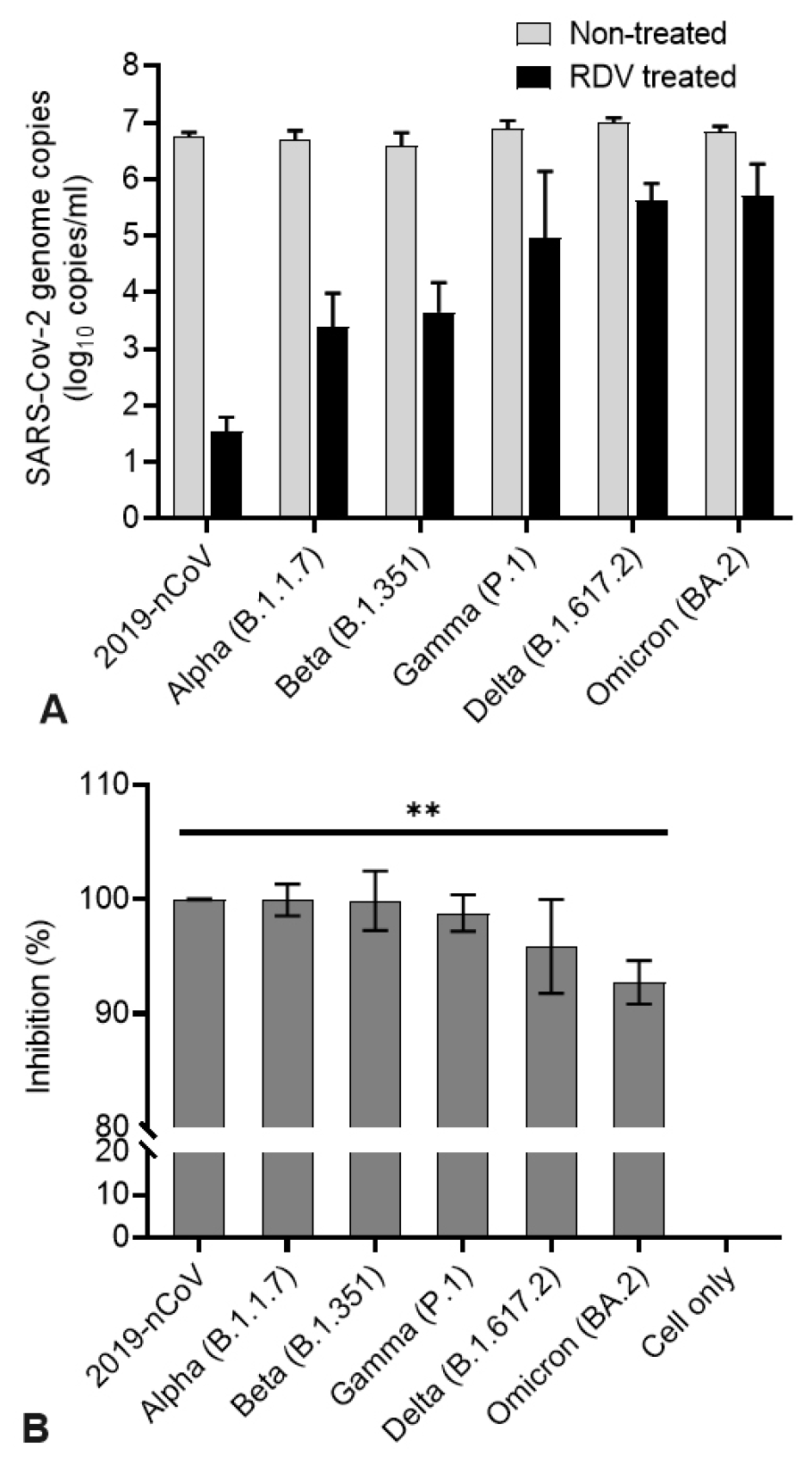 | Fig. 4The antiviral activities of remdesivir (RDV) against SARS-CoV-2 variants. Vero E6 cells were infected with SARS-CoV-2 variants, and after 48 h of RDV treatment, the viral yield of the cell supernatants was quantified by qRT-PCR. (A) Quantifying absolute viral RNA copies (per ml) was determined by qRT-PCR analysis. (B) Inhibitory activity of RDV. Data are presented as mean ± standard deviation of three independent experiments (**p<0.01). 
|
Similar to the results of the plaque reduction and TCID50 analyses, the results of the qRT-PCR analysis confirmed the antiviral activity of RDV. In addition, the antiviral effect of RDV was higher against 2019-nCoV than the other variants.
Go to :

DISCUSSION
In recent years, the development of a fast, sensitive, and comparable method for viral quantification has significantly increased the value of antiviral drug screening and vaccine efficacy testing. Several methods for detecting infectious SARS-CoV-2, including the standard plaque and TCID
50 assays, have been reported and validated
(28,
29,
30,
31). However, this approach has several limitations, such as the biological safety measures involved, stringent laboratory procedures, duration of experiments, and laboratory facilities
(32). To address the absence of standardized testing methods within these limitations, we proposed that an optimized
in vitro assay would be comparable and reliable for SARS-CoV-2 quantification and antiviral efficacy evaluation.
The TCID50 assay counts in this study were approximately ten-fold greater than those of the plaque assay. The mean titer of viruses measured by the TCID50 assay was 8.2 × 106 TCID50/ml, whereas that of the plaque assay was 9.6 × 105 PFU/ml. An approximately ten-fold difference was observed between the counts measured by each assay for the mean and median, indicating that 1 TCID50 is approximately 0.1 PFU for this viral stock under the same conditions. Moreover, the higher counts in the TCID50 assay suggest that a wider range of virus titers can be measured than that by the plaque assay.
It is not unusual for the TCID
50 assay to yield higher titers than the plaque assay; this difference can be attributed to the variation in the pattern of cells detached by plaque or CPE observed in the plate. In the TCID
50 assay, viruses appear as large regions owing to their mobility between cells, whereas in plaque assays, agar overlays are observed as smaller regions as they limit virus migration. Given that the two assays have different times and labor requirements for testing, the TCID
50 assay is considered a better option for a quick realization of test results. Additionally, regarding capacity and range, the TCID
50 assay can test more dilutions with fewer plates than the plaque assay
(33).
In this study, antiviral efficacy was tested using RDV as a direct antiviral agent
(34). The 50% effective dose (EC
50) of RDV against 2019-nCoV was 0.22-0.32 uM. The selectivity index (SI) value of RDV was 312.5-454.5, which can be calculated as the therapeutic index (CC
50 divided by EC
50). The EC
50 values of these RDVs were different from those reported in other studies. A previous study demonstrated that phosphorylated GS-441524, the active molecule of RDV, inhibits SARS-CoV in human airway epithelial cells with an EC
50 value of 0.069 μM
(35), and another demonstrated that RDV had an EC
50 value of 0.77 or 23.15 uM against SARS-CoV-2 in Vero E6 cells
(36). In addition, it was observed that the antiviral effect on SARS-CoV-2 variants of RDV was different, suggesting the need for development of more effective antiviral agents against various viral mutants.
VeroE6 is the preferred cell line for several viral titration assays for SARS-CoV-2. Therefore, although it was used for SARS-CoV-2 titration and evaluation in this study, antiviral effects can be measured differently depending on the cell line. In addition to the cell line concern, despite using the same antiviral agent, different activity values, which may be attributed to different test conditions such as cell line type, virus inoculation amount, drug treatment time point (pre-, post-, or co-treatment), and treatment time, were observed.
RDV is a nucleotide analog prodrug that inhibits SARS-CoV-2 RNA-dependent RNA polymerase (RdRp). Its viral activity against SARS-CoV-2 has been demonstrated in both
in vitro and
in vivo studies
(37). In this study, the antiviral activity of RDV was verified using only
in vitro assays; however, our protocols could be utilized for
in vitro and
in vivo screening of novel therapeutic candidates. The established assay can also be utilized, as identifying and considering the cell- or host-dependent pharmacological actions of drug candidates should be possible when determining drug administration protocols.
Unlike direct-acting antivirals, such as RDV, host-directed therapies target host cell components required for viral replication or attenuate the uncontrolled inflammatory response to infection
(38,
39). Therefore, it is important to test anti-inflammatory responses to develop a therapeutic agent for COVID-19
(40). In addition to cytotoxicity analysis, it is necessary to investigate the potential side effects of RDV on normal cells and its mode of action against SARS-CoV-2. Until recently, the antiviral effect analysis of RDV performed in Vero E6 cells revealed that RDV was ineffective in primary cells or several animal models
(24,
41). Therefore, it is essential to test antiviral drugs
in vivo using infectious animal models to better analyze their antiviral efficacy.
Using the in vitro assays validated in this study, the screening and evaluation of antiviral agents can accelerate and simplify the preclinical drug development process for COVID-19. A streamlined preclinical development pipeline will help to advance the success of therapeutic development.
Go to :









 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download