Dear Editor,
The PICALM-MLLT10 rearrangement (PICALM-MLLT10r) resulting from t(10;11)(p13;q21) can activate the HOXA gene cluster [1, 2], which is considered to be the dominant mechanism underlying leukemic transformation [3, 4]. PICALM-MLLT10r occurs in ~10% of T-cell lymphoblastic leukemia/lymphoma cases and is rarely reported in B-cell lymphoblastic leukemia/lymphoma, mixed-phenotype acute leukemia, lymphoma, and AML [5-7]. We report a case of AML with PICALM-MLLT10r presenting with extensive skin lesions. The Institutional Review Board of Samsung Medical Center, Seoul, Korea approved this report and waived the need for informed consent (2022-06-093).
A 39-year-old man was admitted to our hospital on February 2022 with erythematous patches on his face and trunk. The lesion had appeared on his face three months before, spread to the trunk and upper extremities, and was suspected to have lymphomatous involvement. Positron emission tomography/computed tomography revealed hypermetabolic lesions in the pancreas, anterior mediastinum, and multiple lymph nodes, including the bilateral neck, supraclavicular, axillary, and mediastinal lymph nodes. Initial complete blood count (CBC) was as follows: white blood cells, 16.69×109/L with 75% blasts; Hb, 173 g/L; and platelets, 203×109/L. A bone marrow smear showed 83% blasts with hypercellularity (Fig. 1A–F). Flow cytometry analysis revealed that blasts were positive for CD34, cytoplasmic MPO, CD117, CD33, and HLA-DR; weakly positive for CD7, CD64, and CD123; and negative for cytoplasmic and surface CD3, CD19, CD10, cytoplasmic CD22 and CD79a, CD4, and CD56, which was compatible with AML. Leukemic involvement of the skin lesions on the abdomen, back, and flanks in our patient was confirmed via punch biopsy (Fig. 1G–J).
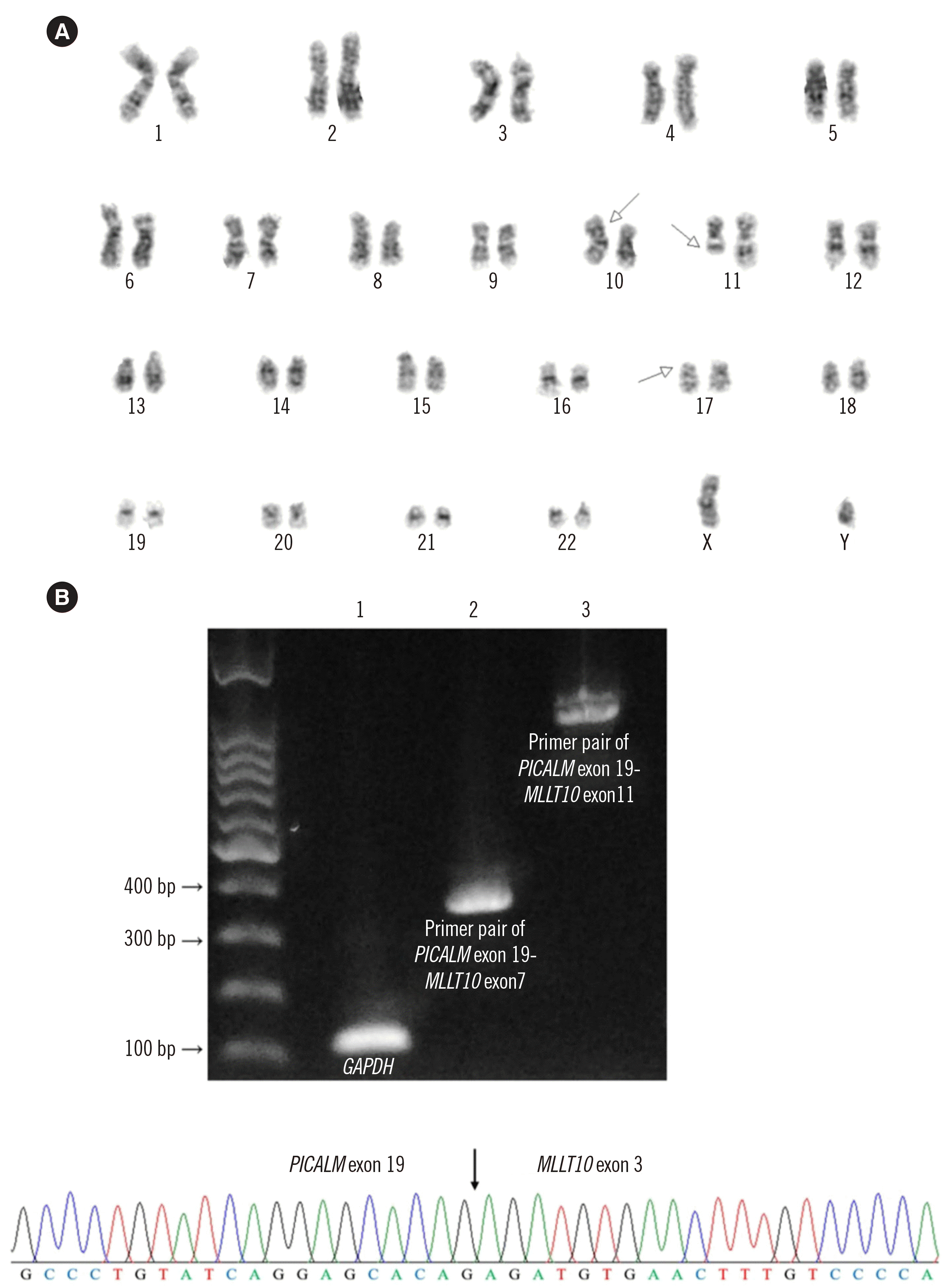
Cytogenetic analysis revealed a complex karyotype: 46,XY,t(10;11)(p13;q21),del(17)(p12)[16]/45,idem,der(12)t(12;17)(p12;q21),-del(17)[4] (Fig. 2A). FISH using the Vysis LSI TP53/CEP17 dual-color probe (Abbott Laboratories, Abbott Park, IL, USA) showed 86.5% of cells with a TP53 deletion. Reverse transcription-PCR for PICALM-MLLT10r was performed using previously reported primers [8]; direct sequencing confirmed the fusion of PICALM exon 19 and MLLT10 exon 3 (Fig. 2B). Targeted next-generation sequencing (NGS) of 46 AML-related genes on NextSeq 550Dx (Illumina, San Diego, CA, USA) identified four mutations: JAK2 p.Gly402Aspfs*9 (NM_004972.3:c.1202_1203insCGACATCATTACCTCTGTAAAGAAGTAGCACCTCCAGCCGTGCTTGAAAATATACAAAGCAACTGTCA; variant allele frequency, 22%), NRAS p.Gly12Ser (NM_002524.4:c.34G>A; 39%), PHF6 p.Cys297Arg (NM_032458.2:c.889T>C; 79%), and TP53 p.Arg248Gln (NM_000546.5:c.743G>A; 67%). After the final diagnosis of AML, the patient received induction chemotherapy with standard-dose cytarabine and idarubicin; however, because of induction failure, re-induction chemotherapy with high-dose cytarabine and idarubicin was administered. During chemotherapy, the skin lesions showed variable improvement and recurrence. After re-induction, the patient developed neutropenic fever and suspected fungal pneumonia and expired three months after the initial diagnosis while planning for allogeneic hematopoietic stem cell transplantation.
AML with PICALM-MLLT10r is very rare and clinically characterized by a young age of onset with frequent extramedullary involvement. To the best of our knowledge, this is the first reported case of AML with PICALM-MLLT10r with extensive skin involvement mimicking cutaneous lymphoma. Skin involvement in hematologic malignancies is mainly observed in primary T-cell lymphomas, such as mycosis fungoides, adult T-cell leukemia/lymphoma, and primary cutaneous B-cell lymphoma, but rarely in AML [9]. In particular, blastic plasmacytoid dendritic cell neoplasm, a rare and aggressive hematologic malignancy that frequently invades the skin and bone marrow [5], was strongly suspected clinically in our patient. However, our patient showed a typical AML immunophenotype, except for ectopic CD7 expression, which is recurrently observed in AML with PICALM-MLLT10r [2, 6].
The prognostic impact of PICALM-MLLT10r in AML is not well established. A few studies have suggested a poor prognosis [2, 6], and our patient indeed showed a very poor prognosis; however, more data are needed. TP53 is a tumor suppressor gene associated with poor prognosis in AML [10]. In our patient, biallelic TP53 inactivation identified by targeted NGS and FISH may have contributed to his poor prognosis. At present, there are no reports on the mutation spectrum of AML with PICALM-MLLT10r, except for one report of a single TP53 mutation in an AML patient with PICALM-MLLT10r that developed after myeloid sarcoma [11]. More data are needed to evaluate the association between TP53 mutations and AML with PICALM-MLLT10r, which may affect patient prognosis. Additionally, we detected a large (68-bp) insertion mutation in JAK2 exon 9 causing a frameshift, which may be related to the erythrocytosis observed in the patient’s initial CBC. However, there was no morphological evidence of myeloproliferative neoplasms in the bone marrow, and the clinical significance of loss-of-function mutations of JAK2 is unclear. In conclusion, this is the first reported case of AML with PICALM-MLLT10r presenting with extensive skin involvement, showing poor prognosis, with concomitant TP53 mutation.
Notes
REFERENCES
1. Dreyling MH, Martinez-Climent JA, Zheng M, Mao J, Rowley JD, Bohlander SK. 1996; The t(10;11)(p13;q14) in the U937 cell line results in the fusion of the AF10 gene and CALM, encoding a new member of the AP-3 clathrin assembly protein family. Proc Natl Acad Sci U S A. 93:4804–9. DOI: 10.1073/pnas.93.10.4804. PMID: 8643484. PMCID: PMC39360.
2. Savage NM, Kota V, Manaloor EJ, Kulharya AS, Pierini V, Mecucci C, et al. 2010; Acute leukemia with PICALM-MLLT10 fusion gene: diagnostic and treatment struggle. Cancer Genet Cytogenet. 202:129–32. DOI: 10.1016/j.cancergencyto.2010.07.126. PMID: 20875875.
3. De Braekeleer E, Douet-Guilbert N, Basinko A, Le Bris MJ, Morel F, De Braekeleer M. 2014; Hox gene dysregulation in acute myeloid leukemia. Future Oncol. 10:475–95. DOI: 10.2217/fon.13.195. PMID: 24559452.
4. Lim HJ, Lee JH, Lee YE, Baek HJ, Kook H, Park JH, et al. 2021; The First Korean Case of NUP98-NSD1 and a Novel SNRK-ETV6 Fusion in a Pediatric Therapy-related Acute Myeloid Leukemia Patient Detected by Targeted RNA Sequencing. Ann Lab Med. 41:443–6. DOI: 10.3343/alm.2021.41.4.443. PMID: 33536367. PMCID: PMC7884187.

5. Swerdlow SH, Campo E, editors. 2017. WHO classification of tumours of haematopoietic and lymphoid tissues. 4th ed. International Agency for Research on Cancer (IARC);Lyon: p. 173–209.
6. Borel C, Dastugue N, Cances-Lauwers V, Mozziconacci MJ, Prebet T, Vey N, et al. 2012; PICALM-MLLT10 acute myeloid leukemia: a French cohort of 18 patients. Leuk Res. 36:1365–9. DOI: 10.1016/j.leukres.2012.07.008. PMID: 22871473.
7. Krzisch D, Zduniak A, Veresezan EL, Daliphard S, Contentin N, Penther D, et al. 2021; Successful treatment of T/myeloid mixed-phenotype acute leukemia with the translocation (10;11)(p13;q14) PICALM/AF10 with 3 + 7 myeloid standard treatment: a case report. Clin Case Rep. 9:1507–13. DOI: 10.1002/ccr3.3815. PMID: 33768878. PMCID: PMC7981630.

8. Laforêt MP, Turlure P, Lippert E, Cornillet-Lefebvre P, Pigneux A, Pradeau R, et al. 2013; Design and feasibility of a novel, rapid, and simple fluorescence 26-plex rt-PCR assay for simultaneous detection of 24 fusion transcripts in adult acute myeloid leukemia. J Mol Diagn. 15:186–95. DOI: 10.1016/j.jmoldx.2012.11.004. PMID: 23318495.

9. Han JH, Ko YH, Kang YK, Kim WS, Kim YJ, Kim I, et al. 2014; Characteristics of cutaneous lymphomas in Korea according to the New WHO-EORTC classification: report of a nationwide study. Korean J Pathol. 48:126–32. DOI: 10.4132/KoreanJPathol.2014.48.2.126. PMID: 24868225. PMCID: PMC4026803.

10. Short NJ, Montalban-Bravo G, Hwang H, Ning J, Franquiz MJ, Kanagal-Shamanna R, et al. 2020; Prognostic and therapeutic impacts of mutant TP53 variant allelic frequency in newly diagnosed acute myeloid leukemia. Blood Adv. 4:5681–9. DOI: 10.1182/bloodadvances.2020003120. PMID: 33211826. PMCID: PMC7686900.
11. Naesens L, Devos H, Nollet F, Michaux L, Selleslag D. 2018; Mediastinal myeloid sarcoma with TP53 mutation preceding acute myeloid leukemia with a PICALM-MLLT10 fusion gene. Acta Haematol. 140:97–104. DOI: 10.1159/000491596. PMID: 30227397.
Fig. 1
Bone marrow (BM) aspiration and biopsy. Medium-to-large size leukemic blasts were observed in (A) BM aspiration (Wright-Giemsa stain, ×400) and (B) biopsy (hematoxylin and eosin stain, ×100). In leukemic blasts, (C) myeloperoxidase (MPO) were positive, (D) CD3 were negative, (E) CD34 were positive, and (F) terminal deoxynucleotidyl transferase (TdT) immunostains (×100) were partially positive. (G-J) Skin biopsy of trunk. (G) Leukemic blasts were observed (hematoxylin and eosin stain, ×100); (H) MPO was positive (×400) and (I) CD3 and (J) CD20 were negative (×100).

Fig. 2
Cytogenetic and molecular analysis of bone marrow study (A) Chromosomal analysis showed 46,XY,t(10;11)(p13;q21),del(17)(p12). Each arrow indicates the reciprocal translocation between 10p13 and 11q21, and deletion of 17p. (B) The PICAM-MLLT10 fusion transcript was identified via agarose gel electrophoresis (Lane 1, internal control using GAPDH [amplicon size, 131 bp]; lanes 2 and 3, PICALM-MLLT10 fusion transcripts identified using the PICALM exon 19 and MLLT10 exon 7 primer pair 1 [358 bp] and PICALM exon 19 and MLLT10 exon 11 primer pair 2 [922 bp], respectively) and Sanger sequencing. Chromatogram of Sanger sequencing shows the PICALM exon 19-MLLT10 exon 3 fusion.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download