Abstract
Background
Comparative analysis of virulence factors (VFs) between Pasteurella canis and Pasteurella multocida are lacking, although both cause zoonotic infections. We determined the virulence-associated genome sequence characteristics of P. canis and assessed the toxin gene prevalence unique to P. canis among clinical isolates of P. canis and P. multocida.
Methods
We selected 10 P. canis and 16 P. multocida whole-genome sequences (WGSs) from the National Center for Biotechnology database. The VFanalyzer tool was used to estimate P. canis-characteristic VFs. Amino acid sequences of VFs were compared with multiple-aligned sequences. The genome structure containing P. canis-characteristic and adjacent loci was compared to the corresponding P. multocida genome structure. After designing primer sequences and assessing their accuracy, we examined the gene prevalence of the P. canis-characteristic VFs using PCR among clinical isolates of P. multocida and P. canis.
Results
Using VFanalyzer, we found virulence-associated cytolethal distending toxin (cdt)A–cdtB–cdtC loci common to all P. canis WGSs that were not found in P. multocida WGSs. Similarities in the multiple alignments of CdtA–CdtB–CdtC amino acid sequences were found among the 10 P. canis WGSs. Shared or similar loci around cdtA–cdtB–cdtC were identified between the P. canis and P. multocida genome structures. The PCR-based cdtA–cdtB–cdtC prevalence differed for P. canis and P. multocida clinical isolates.
Pasteurella multocida was first isolated by Louis Pasteur in 1881 from samples received during an epidemiological survey of a fowl cholera outbreak. Pasteurella canis was previously classified as P. multocida biotype 6 or “dog type” and was then reclassified in 1985 based on DNA homology [1]. P. canis is a tiny, non-motile, facultatively anaerobic, gram-negative coccobacillus belonging to a species of G-Proteobacteria in the family Pasteurellaceae. P. canis forms smaller colonies as against P. multocida that forms larger colonies when the isolates are inoculated onto blood agar plates and incubated in 5% CO2 at 37°C for 24 hours, suggesting that P. canis has relatively slower growth capacity than P. multocida under the same culture conditions [2]. With reclassification, P. multocida was divided into three subspecies: P. multocida subsp. multocida, P. multocida subsp. septica, and P. multocida subsp. gallicida [1]. Phylogenetic trees showing relationships between 16S rRNA sequences and fragment sequences (449–473 bp) of the sodA gene from type strains of Pasteurella demonstrated that P. canis is more closely related to P. stomatis and P. dagmatis than to P. multocida subspp. multocida, septica, and gallicida [3]. With respect to the biochemical properties of P. canis, the ornithine decarboxylation property is positive, whereas the urease test is negative, and variable reactions are obtained for fermentation of trehalose and D-xylose [1]. Biotype 1 isolates are positive for indole formation, whereas biotype 2 isolates are negative for indole formation.
P. canis biotype 1 is mainly observed in the oral cavities of dogs and is also often isolated from the wound sites of humans after receiving dog bites [1], whereas biotype 2 isolates are mainly recovered from calves. There are several reports of P. canis-infected humans with bacteremia [4], soft tissue infection [5], eye infection [6], respiratory infection [7], septic arthritis [8], osteomyelitis [9], gastrointestinal infection [10], breast implant infection [11], and peritonitis [12]. The Emergency Medicine Animal Bite Infection Study Group [13] performed a bacteriological assay of infected wound sites resulting from dog and cat bites, showing that P. canis was the most common species isolated from wounds caused by dog bites, whereas P. multocida subspp. multocida and septica were the most common bacteria isolated from wounds caused by cat bites. Basic investigations of P. canis isolates are scarce, especially studies on its virulence factors (VFs).
The whole-genome sequence (WGS) of a blood-origin P. canis isolate from a diseased male Saint-Bernard dog in Japan was recently reported [2]. No comparative genomic analysis of VFs between P. canis and P. multocida has been performed to date, although both cause zoonotic infections. This study aimed to compare the genome sequence characteristics of P. canis and P. multocida using the VF-detection tool VFanalyzer, which provides an automatic analysis pipeline (http://www.mgc.ac.cn/cgi-bin/VFs/v5/main.cgi?func=VFanalyzer) for the systematic screening of known or potential VFs in complete or draft bacterial genomes [14, 15]. We also assessed the prevalence of each unique VF gene for the two species (P. canis and P. multocida) among clinical isolates.
We extracted WGSs from P. canis (N=10) and P. multocida (N=16) deposited in the National Center for Biotechnology Information (NCBI) database (updated June 3, 2022) for this retrospective study. Sixteen P. multocida WGSs were selected covering various sequence types (including newly identified sequence types), demonstrating the high genomic diversity of this species. Among the 10 P. canis WGSs, seven are newly added, including three human isolates (two from Korea and one from Japan) and four dog isolates (three from Korea and one from Japan). Table 1 lists the characteristics of the P. canis and P. multocida WGSs (including the strain, host, isolation source, assembly level, collection date/geographic location, and GenBank accession number). Information on the host, isolation source, collection date, and geographic location was retrieved from the corresponding BioSample website or datasheets of collection companies. The included reference WGSs were P. canis National Collection of Type Cultures (NCTC) 11621(T), P. multocida subsp. multocida American Type Culture Collection (ATCC) 43137(T), P. multocida subsp. septica NCTC 11995(T), and P. multocida subsp. gallicida NCTC 10204(T).
To compare the VFs of P. canis and P. multocida, the VFanalyzer tool from the VF database (http://www.mgc.ac.cn/VFs/) was used. Any publicly available bacterial genome can be used as input for VFanalyzer, either from the pre-downloaded genome list or via any valid GenBank accession number. The genus Haemophilus was selected for the analysis because the option of Pasteurella was not available for selection. Each WGS file (complete or draft genome in GenBank format) from P. canis (N=10) and P. multocida (N=16) was entered into VFanalyzer, and the data of known or potential VFs, related genes, and corresponding loci (along with their nucleotide positions/sequences) for each WGS were output. Putative P. canis-characteristic VFs that were not present among the P. multocida-characteristic VFs were estimated. All analyses were conducted at Ōmura Satoshi Memorial Institute, Kitasato University.
To evaluate the uniqueness of the estimated VFs, the corresponding nucleotide/amino acid (AA) sequences were inserted into the NCBI nucleotide/protein–nucleotide Basic Local Alignment Search Tool (BLASTn/BLASTp; https://blast.ncbi.nlm.nih.gov/Blast.cgi). To assess the percent similarities among deduced AA sequences obtained from the nucleotide sequences, multiple alignments were performed using the web-based tool ClustalW (https://www.genome.jp/tools-bin/clustalw).
The complete circular genomic structure was constructed for P. canis HL_NV12211 (CP085871.1) as a representative, based on the WGS data obtained in this study, which is now recommended in the NCBI database as the P. canis reference genome (https://www.ncbi.nlm.nih.gov/data-hub/taxonomy/753/) (Table 1). This isolate was recovered from a Korean canine pus sample (Table 1). The WGS from isolate ATCC 43137(T) (CP008918.1) was used as a representative P. multocida subsp. multocida genome (Table 1) to construct the comparative genomic structure. The gene information on HL_NV12211 and ATCC 43137(T) WGSs was obtained from GenBank in the NCBI database, including the directions and lengths of genes, locus tags, and gene–gene distances. The prokaryotic genome annotation pipeline was used as a tool for annotation revision. We assessed the shared, similar, and unique loci between HL_NV12211 and ATCC 43137(T) WGSs.
We designed PCR primer sets (forward and reverse) using the web-based Primer3Plus tool (https://www.primer3plus.com). The specificity of the primer sequences was examined by inserting them into NCBI BLASTn. To assess the accuracy of the primer sequences, we conducted simulated PCR assays using Serial Cloner 2.6 (http://serialbasics.free.fr/Serial_Cloner.html) based on the obtained WGSs [16, 17].
Companion animal- and human-origin isolates from Japan were identified from PCR-based 16S rRNA sequencing data [18]. Thirty P. canis isolates from dogs (N=16), a cat (N=1), and humans (N=13) and 48 P. multocida isolates from dogs (N=5), cats (N=27), and humans (N=16) were included for this analysis. Isolation sources were mainly the pus/skin (N=44), upper/lower respiratory tract (N=25), and ear (N=6), along with the eye, blood, or pleural effusion (N=1 each). The collection years were mainly 2015–2019 (N=62) and 2021 (N=12), along with 1997, 1998, 2010, and 2013 (N=1 each). The geographic locations (prefectures in Japan) were mainly Chiba (N=40), Tokyo (N=15), Ibaraki (N=6), Saitama (N=5), Kanagawa (N=3), Aichi (N=3), Gifu (N=2), and Nagasaki (N=2), along with Okayama and Fukuoka (N=1 each).
We used primer sets for PCR amplification that were identical to the simulation PCR-based primer sets. One isolate (either PA42 or PA57) was used as a positive control (Table 1), and DNase/RNase/protease-free water was used as a negative control in each PCR assay. PCR products were examined by 1.5% agarose gel electrophoresis in Tris-acetate–EDTA buffer. Direct sequencing of several PCR-positive products was performed on Applied Biosystems 3730xl DNA Analyzer with BigDye Terminator V3.1 (Thermo Fisher Scientific, Waltham, MA, USA). The amplification primers were used for the sequencing reaction. The prevalence of each estimated P. canis-unique VF gene was evaluated in all clinical isolates.
The following ethics committees reviewed and approved our study design to maintain the privacy of humans and companion animals: Kitasato Institute Hospital (Tokyo, Japan; approval no. 21061), Kitasato University Medical Center (Saitama, Japan; approval no. 2021033), Sanritsu Zelkova Veterinary Laboratory (Tokyo, Japan; approval no. SZ20220525), Sanritsu Laboratory (Chiba, Japan; approval no. 22-01), and Chiba Kaihin Municipal Hospital (Chiba, Japan; approval no. 2021-02).
Background information (host species, isolation source, collection date, and geographic location) for the selected WGSs is publicly available from the online NCBI database.
Fisher’s exact test (two-sided) was performed to analyze the significance of the association of the prevalence of each unique VF gene between P. canis and P. multocida. The associations of isolation sources (pus/skin, upper/lower respiratory tract, and ear) with host (animals, N=49; humans, N=29) or isolate (P. canis, N=30; P. multocida, N=48) were also determined using Fisher’s exact test (two-sided). Statcel4 (OMS Publisher, Tokyo, Japan) was used for statistical analysis. P<0.05 indicated statistical significance.
The VFs obtained from VFanalyzer in querying the Haemophilus genus included those related to adherence (e.g., HMW1, HMW2, hemagglutinating pili), endotoxin (lipooligosaccharide), immune evasion (e.g., exopolysaccharide, IgA1 protease, P2 porin), iron uptake (e.g., Haemophilus iron transport locus, heme biosynthesis, heme/hemopexin-binding complex), toxins (cytolethal distending toxin [Cdt] and hemolysin), cell-surface components (trehalose-recycling ABC transporter), nutritional virulence (pyrimidine biosynthesis from Francisella), secretion system (Hcp secretion island-1-encoded type VI secretion system [H1-T6SS] and Legionella vir homologs [Lvh] type IVa secretion system), stress adaptation (catalase from Neisseria and SodCI from Salmonella), and antiphagocytosis (capsular polysaccharide from Vibrio and capsule from Klebsiella). The prevalence and composition of VFs, related genes, and corresponding loci varied between the P. canis and P. multocida genomes. We identified cdtA–cdtB–cdtC sequences common to the 10 P. canis WGSs, which were absent among the 16 P. multocida WGSs; thus, the cdtA–cdtB–cdtC sequences were considered potential P. canis-characteristic VFs. The HL_NV12211 WGS included a cdtA locus of 756 bp (nucleotide positions 2,095,545 to 2,096,300), a cdtB locus of 843 bp (nucleotides 2,096,316 to 2,097,158), and a cdtC locus of 543 bp (nucleotides 2,097,169 to 2,097,711).
Multiple nucleotide/AA sequence alignment of the cdtA–cdtB–cdtC loci of the P. canis HL_NV12211 WGS showed high sequence similarities among P. canis populations but not among populations of P. multocida or other Pasteurella species. Fig. 1 shows the multiple AA sequence alignments of CdtA–CdtB–CdtC loci. High similarities (percentages) of deduced AA sequences were found between HL_NV12211 CdtA (251 AA), CdtB (280 AA), and CdtC (180 AA) and the sequences of the other P. canis genomes, with the lowest similarities of 98.8%, 98.6%, and 98.9%, respectively.
Fig. 2 shows the genome structure containing the cdtA–cdtB–cdtC loci and adjacent loci in the P. canis HL_NV12211 WGS. Compared to that in the ATCC 43137(T) WGS, shared loci (relA, recO, rsmE, ruvX/DR93_65, eno, and pyrG), similar loci (rlmD/rumA with AA sequence similarity of 67.3% and K7G93_001967/DR93_66 with AA sequence similarity of 89.2%), and loci were identified in the P. canis WGS (cdtA–cdtB–cdtC and K7G93_001965) but not in the P. multocida WGS. The loci K7G93_001965, K7G93_001967, and DR93_66 are predicted to encode hypothetical proteins.
Primer sequences are located within the corresponding open reading frames to amplify sequences with expected sizes of 693 bp, 582 bp, and 433 bp. Table 2 shows the oligonucleotide primers and their corresponding melting temperature (Tm) values. PCR was performed with 30 cycles consisting of denaturation at 98°C for 10 seconds, annealing at 52°C for 30 seconds, and extension at 72°C for 45 seconds to amplify cdtA; 30 cycles consisting of denaturation at 98°C for 10 seconds, annealing at 53°C for 30 seconds, and extension at 72°C for 40 seconds to amplify cdtB; and 30 cycles consisting of denaturation at 98°C for 10 seconds, annealing at 52°C for 30 seconds, and extension at 72°C for 30 seconds to amplify cdtC.
We found no associations between the isolation sources and host (animals/humans) or pathogen species (P. canis/P. multocida): pus/skin vs. host/pathogen species P=0.102/P=1.0, respiratory tract vs. host/pathogen species P=1.0/P=0.619, and ear vs. host/pathogen species P=0.079/P=0.397.
Fig. 3 shows gel electrophoresis images of the amplified cdtA–cdtB–cdtC products using DNA extracted from the clinical isolates. Table 3 shows the prevalence of each cdt gene in P. canis and P. multocida isolates. High similarities (≥98.6%) were found between cdtA and cdtB–cdtC sequences using several PCR-positive products (N=11 for each gene) and reported HL_NV12211 cdtA–cdtB–cdtC sequences. The nucleotide sequences obtained by PCR amplification and direct sequencing in 11 isolates have been deposited in DDBJ/EMBL/GenBank under accession numbers LC716775–LC716807.
The prevalence of cdtA–cdtB–cdtC was significantly associated with P. canis rather than with P. multocida (P<0.01).
We conducted a challenging study to find virulence-associated genome sequences specific to P. canis. Based on the “One Health” concept [19], which is a comprehensive health control strategy for humans, contact animals, and related environments, bacterial pathogens with VFs that may be circulating should be carefully monitored to maintain an environment of total health. Using VFanalyzer, we identified cdtA–cdtB–cdtC loci as potential P. canis-specific VFs, which were not identified in the WGSs of P. dagmatis NCTC 11617(T) (LT906448.1), P. testudinis NCTC 12150(T) (UGSY00000000.1), P. skyensis 95A1 (type strain) (CP016180.1), P. bettyae NCTC 10535(T) (UFRH00000000.1), and P. bettyae CCUG 2042 (AJSX00000000.1). These three loci may be unique toxin genes and promising targets for the rapid identification of P. canis in clinical settings.
Based on a search of the keywords “pasteurella canis, cytolethal distending toxin” or “pasteurella canis, cdt” in the PubMed database (https://pubmed.ncbi.nlm.nih.gov/), there were no hits for related manuscripts as of September 3, 2022. However, Fukushima, et al. [20] reported that P. canis isolates from the dental plaques of dogs with periodontal disease harbor the cdtA–cdtB–cdtC loci. Thus, our study provides a rare description of cdt genes in P. canis isolates from humans and companion animals in Korea and Japan.
Cdt was discovered in Escherichia coli in 1987 [21], and similar toxin activities have been documented in other enteric pathogens, including Shigella spp. and Campylobacter spp. [21, 22]. Cdt can modulate the host cell cycle by suppressing the G2/M transition. Bacterial genotoxins trigger single-strand and double-strand breaks in DNA of eukaryotic cells and are functionally homologous to mammalian DNase I, leading to alterations in the DNA damage response [23], resulting in cell aging, apoptosis, and genomic instability, which contribute to tumor initiation and progression. Three loci within an operon were identified: cdtA–cdtB–cdtC [24]. Among the three proteins encoded by cdtA–cdtB–cdtC, CdtB was shown to possess nuclease activity [25]. CdtA–CdtC is required for delivering CdtB into host cells, allowing CdtB to translocate to the nucleus, resulting in DNA damage [26]. CdtB nuclease activity in eukaryotic cells can induce an altered DNA damage response, which promotes genomic instability, disturbs the cell cycle, and establishes a chronic pro-inflammatory environment in the host [27].
Using BLASTp on the NCBI web server, we examined similarities in CdtA–CdtB–CdtC AA sequences between isolate HL_NV12211 and other species. The CdtA AA sequence was similar to that of Aggregatibacter actinomycetemcomitans strain UP14 (220 AA and 66.5% similarity) and Haemophilus ducreyi strain 35000HP (223 AA and 65.1% similarity); CdtB was similar to A. actinomycetemcomitans NCTC_9710(T) (283 AA and 90.5% similarity) and H. ducreyi 35000HP (283 AA and 90.5% similarity); and CdtC was similar to H. ducreyi 35000HP (186 AA and 73.6% similarity) and A. actinomycetemcomitans strain RhAa3 (180 AA and 75.0% similarity). The family Pasteurellaceae mainly consists of the genera Pasteurella, Haemophilus, Aggregatibacter, and Actinobacillus as human pathogenic microorganisms. Thus, P. canis CdtA–CdtB–CdtC AA sequences are homologous to those of other species in the family Pasteurellaceae. For example, A. actinomycetemcomitans, a periodontal pathogen colonizing the human oral cavity, possesses CdtA–CdtB–CdtC as an exotoxin [28]. The possibility that P. canis is related to periodontal disease in dogs and cats has been reported [20]. A functional study of H. ducreyi CdtB and the endocytosis of CdtA dissociated from H. ducreyi CdtB–CdtC were recently reported [29, 30]. Further biological and functional analyses of P. canis CdtA–CdtB–CdtC are needed.
This study has two main limitations. First, we included five complete and five contig WGSs. We cannot deny the possibility that other characteristic genome sequences may be located within the gapped regions of contig WGSs. To enhance the quality of comparative genome analysis, the complete WGSs of P. canis should be determined and uploaded to the NCBI database. Second, we obtained limited information on host demographics (host species, isolation source, collection date, and geographic location). Detailed information on the underlying situation, diagnosis of infections, therapeutic strategies, and outcomes need to be collected from medical doctors and veterinarians.
Our observations suggest the prevalence of cdtA–cdtB–cdtC genes (as potential VFs) as a unique characteristic of P. canis isolates from humans and companion animals in Japan. Fukushima, et al. [20] found that supernatants from bacterial lysis had toxic and lethal effects on HeLa cells. We also plan to demonstrate the presence of CdtA–CdtB–CdtC within the supernatants of the isolate broth cultures and to evaluate the toxic and lethal effects on HeLa cells and other human cell lines. Future studies are also needed to monitor variations in cdtA–cdtB–cdtC prevalence among similar populations. The clinical significance of CdtA–CdtB–CdtC in humans and companion animals should be clarified to facilitate interpretation and clinical decision-making for medical doctors and veterinarians.
ACKNOWLEDGMENTS
We thank Goro Kurita, D.M.V. (Laboratory of Infectious Diseases, Graduate School of Infection Control Sciences, Kitasato University, Tokyo, Japan), Prof. Noriyuki Nagano (Department of Medical Sciences, Graduate School of Medicine, Science and Technology, Shinshu University, Nagano, Japan), and Mr. Tomohiro Fujita (Department of Clinical Laboratory, Kitasato University Medical Center, Kitasato University, Saitama, Japan) for their assistance.
Notes
AUTHOR CONTRIBUTIONS
Conceptualization: Yoshida H and Takahashi T; Investigation: Yoshida H; Formal Analysis: Yoshida H, Kim J-M, Maeda T, Goto M, Kim J-S, and Okuzumi K; Resources: Tsuyuki Y, Shibata S, Shizuno K, Okuzumi K, and Takahashi T; Writing — Original Draft Preparation: Takahashi T; Writing — Review and Editing: Yoshida H.
REFERENCES
1. Mutters R, Ihm P, Pohl S, Frederiksen W, Mannheim W. 1985; Reclassification of the genus Pasteurella Trevisan 1887 on the basis of deoxyribonucleic acid homology, with proposals for the new species Pasteurella dagmatis, Pasteurella canis, Pasteurella stomatis, Pasteurella anatis, and Pasteurella langaa. Int J Syst Bacteriol. 35:309–22. DOI: 10.1099/00207713-35-3-309.
2. Maeda T, Yoshida H, Kim JM, Tsuyuki Y, Kurita G, Kim JS, et al. 2022; Draft genome sequence of blood-origin Pasteurella canis strain PA42, isolated from a dog in Japan. Microbiol Resour Announc. 11:e0026022. DOI: 10.1128/mra.00260-22. PMID: 35638811. PMCID: PMC9302189.

3. Gautier AL, Dubois D, Escande F, Avril JL, Trieu-Cuot P, Gaillot O. 2005; Rapid and accurate identification of human isolates of Pasteurella and related species by sequencing the sodA gene. J Clin Microbiol. 43:2307–14. DOI: 10.1128/JCM.43.5.2307-2314.2005. PMID: 15872260. PMCID: PMC1153776.

4. Albert TJ, Stevens DL. 2010; The first case of Pasteurella canis bacteremia: a cirrhotic patient with an open leg wound. Infection. 38:483–5. DOI: 10.1007/s15010-010-0040-1. PMID: 20623245.

5. Kim B, Pai H, Lee KH, Lee Y. 2016; Identification of Pasteurella canis in a soft tissue infection caused by a dog bite: the first report in Korea. Ann Lab Med. 36:617–9. DOI: 10.3343/alm.2016.36.6.617. PMID: 27578520. PMCID: PMC5011120.

6. Shah A, Talati M, Mauger T. 2017; Medical and surgical management of Pasteurella canis infectious keratitis. IDCases. 9:42–4. DOI: 10.1016/j.idcr.2017.05.012. PMID: 28660128. PMCID: PMC5479940.

7. Bhat S, Acharya PR, Biranthabail D, Rangnekar A, Shiragavi S. 2015; A case of lower respiratory tract infection with canine-associated Pasteurella canis in a patient with chronic obstructive pulmonary disease. J Clin Diagn Res. 9:DD03–4. DOI: 10.7860/JCDR/2015/13900.6351. PMID: 26435948. PMCID: PMC4576539.
8. Hazelton BJ, Axt MW, Jones CA. 2013; Pasteurella canis osteoarticular infections in childhood: review of bone and joint infections due to pasteurella species over 10 years at a tertiary pediatric hospital and in the literature. J Pediatr Orthop. 33:e34–8. DOI: 10.1097/BPO.0b013e318287ffe6. PMID: 23482278.
9. Hara H, Ochiai T, Morishima T, Arashima Y, Kumasaka K, Kawano KY. 2002; Pasteurella canis osteomyelitis and cutaneous abscess after a domestic dog bite. J Am Acad Dermatol. 46(S5):S151–2. DOI: 10.1067/mjd.2002.106350. PMID: 12004298.
10. Mensah-Glanowska P, Fornagiel S, Chrzan R, Ulatowska-Białas M, Piątkowska-Jakubas B. 2020; Of horses and zebras: a gastrointestinal infection with Pasteurella canis in a patient with acute myeloid leukemia. Pol Arch Intern Med. 130:335–7. DOI: 10.20452/pamw.15142. PMID: 31933489.
11. Hannouille J, Belgrado JP, Vankerchove S, Vandermeeren L. 2019; Breast implant infection with Pasteurella canis: first case-report. JPRAS Open. 21:86–8. DOI: 10.1016/j.jpra.2019.07.006. PMID: 32158890. PMCID: PMC7061584.

12. Castellano I, Marín JP, Gallego S, Mora M, Rangel G, Suarez MA, et al. 2011; Pasteurella canis peritonitis in a peritoneal dialysis patient. Perit Dial Int. 31:503–4. DOI: 10.3747/pdi.2011.00007. PMID: 21799062.

13. Talan DA, Citron DM, Abrahamian FM, Moran GJ, Goldstein EJ. 1999; Bacteriologic analysis of infected dog and cat bites. Emergency Medicine Animal Bite Infection Study Group. N Engl J Med. 340:85–92. DOI: 10.1056/NEJM199901143400202. PMID: 9887159.

14. Liu B, Zheng D, Jin Q, Chen L, Yang J. VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019; 47:D687–92. DOI: 10.1093/nar/gky1080. PMID: 30395255. PMCID: PMC6324032.

15. Liu B, Zheng D, Zhou S, Chen L, Yang J. VFDB 2022: a general classification scheme for bacterial virulence factors. Nucleic Acids Res. 2022; 50:D912–7. DOI: 10.1093/nar/gkab1107. PMID: 34850947. PMCID: PMC8728188.

16. Takahashi T, Maeda T, Lee S, Lee DH, Kim S. 2020; Clonal distribution of clindamycin-resistant erythromycin-susceptible (CRES) Streptococcus agalactiae in Korea based on whole genome sequences. Ann Lab Med. 40:370–81. DOI: 10.3343/alm.2020.40.5.370. PMID: 32311850. PMCID: PMC7169627.

17. Shin H, Takahashi T, Lee S, Choi EH, Maeda T, Fukushima Y, et al. 2022; Comparing genomic characteristics of Streptococcus pyogenes associated with invasiveness over a 20-year period in Korea. Ann Lab Med. 42:438–46. DOI: 10.3343/alm.2022.42.4.438. PMID: 35177564. PMCID: PMC8859563.

18. Stackebrandt E, Ebers J. 2006; Taxonomic parameters revisited: tarnished gold standards. Microbiol Today. 33:152–5.
19. Centers for Disease Control and Prevention. One Health. https://www.cdc.gov/onehealth/index.html. Updated on Oct 2022.
20. Fukushima M, Hinenoya A, Asakura M, Nishimura K, Hasegawa T, Honda Y, et al. Cell toxic activity of cytolethal distending toxin (cdt) gene-positive Pasteurella canis isolated from oral cavities of dogs. Abstract at the 59th Annual Meeting of the Japanese Society of Bacteriology, Kansai Branch in 2006 (in Japanese).
21. Johnson WM, Lior H. 1987; Production of Shiga toxin and a cytolethal distending toxin (CLDT) by serogroups of Shigella spp. FEMS Microbiol Lett. 48:235–8. DOI: 10.1111/j.1574-6968.1987.tb02548.x.
22. Johnson WM, Lior H. 1988; A new heat-labile cytolethal distending toxin (CLDT) produced by Campylobacter spp. Microb Pathog. 4:115–26. DOI: 10.1016/0882-4010(88)90053-8. PMID: 2849028.
23. Guerra L, Guidi R, Frisan T. 2011; Do bacterial genotoxins contribute to chronic inflammation, genomic instability and tumor progression? FEBS J. 278:4577–88. DOI: 10.1111/j.1742-4658.2011.08125.x. PMID: 21585655.

24. Scott DA, Kaper JB. 1994; Cloning and sequencing of the genes encoding Escherichia coli cytolethal distending toxin. Infect Immun. 62:244–51. DOI: 10.1128/iai.62.1.244-251.1994. PMID: 8262635. PMCID: PMC186093.

25. Elwell CA, Dreyfus LA. 2000; DNase I homologous residues in CdtB are critical for cytolethal distending toxin-mediated cell cycle arrest. Mol Microbiol. 37:952–63. DOI: 10.1046/j.1365-2958.2000.02070.x. PMID: 10972814.

26. Cortes-Bratti X, Chaves-Olarte E, Lagergård T, Thelestam M. 2000; Cellular internalization of cytolethal distending toxin from Haemophilus ducreyi. Infect Immun. 68:6903–11. DOI: 10.1128/IAI.68.12.6903-6911.2000. PMID: 11083812. PMCID: PMC97797.

27. Guidi R, Guerra L, Levi L, Stenerlöw B, Fox JG, Josenhans C, et al. 2013; Chronic exposure to the cytolethal distending toxins of Gram-negative bacteria promotes genomic instability and altered DNA damage response. Cell Microbiol. 15:98–113. DOI: 10.1111/cmi.12034. PMID: 22998585. PMCID: PMC4136655.

28. Belibasakis GN, Maula T, Bao K, Lindholm M, Bostanci N, Oscarsson J, et al. 2019; Virulence and pathogenicity properties of Aggregatibacter actinomycetemcomitans. Pathogens. 8:222. DOI: 10.3390/pathogens8040222. PMID: 31698835. PMCID: PMC6963787.

29. Pons BJ, Loiseau N, Hashim S, Tadrist S, Mirey G, Vignard J. 2020; Functional study of Haemophilus ducreyi cytolethal distending toxin subunit B. Toxins. 12:530. DOI: 10.3390/toxins12090530. PMID: 32825080. PMCID: PMC7551728.

30. Robb Huhn G 3rd, Torres-Mangual N, Clore J, Cilenti L, Frisan T, Teter K. 2021; Endocytosis of the CdtA subunit from the Haemophilus ducreyi cytolethal distending toxin. Cell Microbiol. 23:e13380. DOI: 10.1111/cmi.13380. PMID: 34292647.

Fig. 1
Multiple amino acid (AA) sequence alignments of cytolethal distending toxin (Cdt)A, B, and C between HL_NV12211 (harboring CdtA–CdtB–CdtC of 251 AAs, 280 AAs, and 180 AAs, respectively) and the remaining nine whole-genome sequences using ClustalW version 2.1. “*” indicates positions having a single, fully conserved residue; “:” indicates that one of the following “strong” groups (STA/NEQK/NHQK/NDEQ/QHRK/MILV/MILF/HY/FYW) is fully conserved; and “.” indicates that one of the following “weaker” groups (CSA/ATV/SAG/STNK/STPA/SGND/SNDEQK/NDEQHK/NEQHRK/FVLIM/HFY) is fully conserved. Gray shading shows AA substitutions compared to the consensus sequence of HL_NV12211.
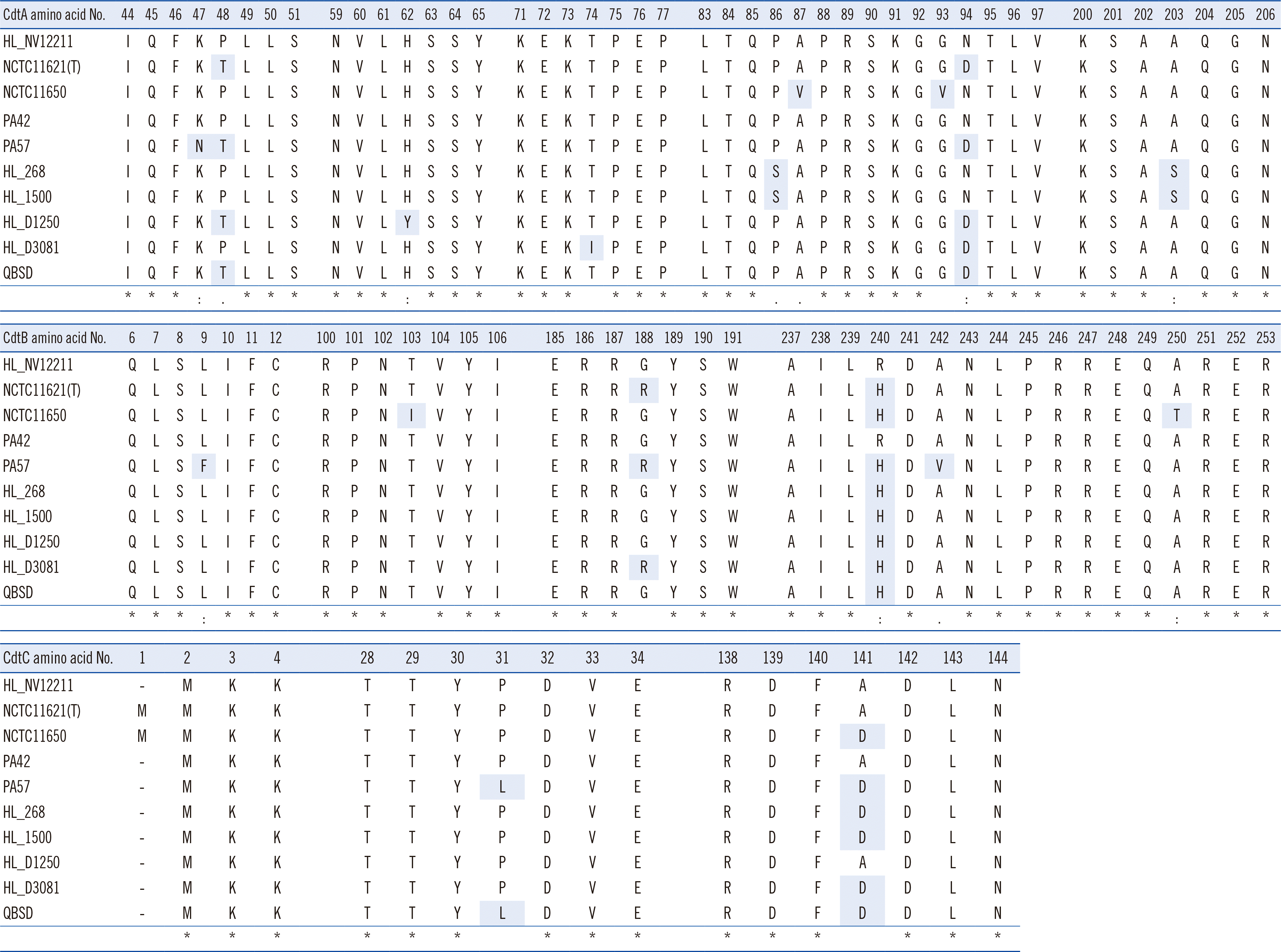
Fig. 2
Genome structure containing the cytolethal distending toxin (cdt)A–cdtB–cdtC loci of Pasteurella canis and adjacent loci from strain HL_NV12211 (GenBank accession no. CP085871.1) (upper) and the comparative structure from Pasteurella multocida subsp. multocida ATCC 43137(T) (GenBank accession no. CP008918.1) (lower). Asterisks show putative Holliday junction resolvase. K7G93_001965, K7G93_001967, and DR93_66 represent loci encoding hypothetical proteins.
Abbreviations: relA, GTP diphosphokinase; rlmD and rumA, 23S rRNA (uracil(1939)-C(5))-methyltransferase; recO, DNA repair protein; rsmE, 16S rRNA (uracil(1498)-N(3))-methyltransferase; eno, phosphopyruvate hydratase; pyrG, CTP synthase.
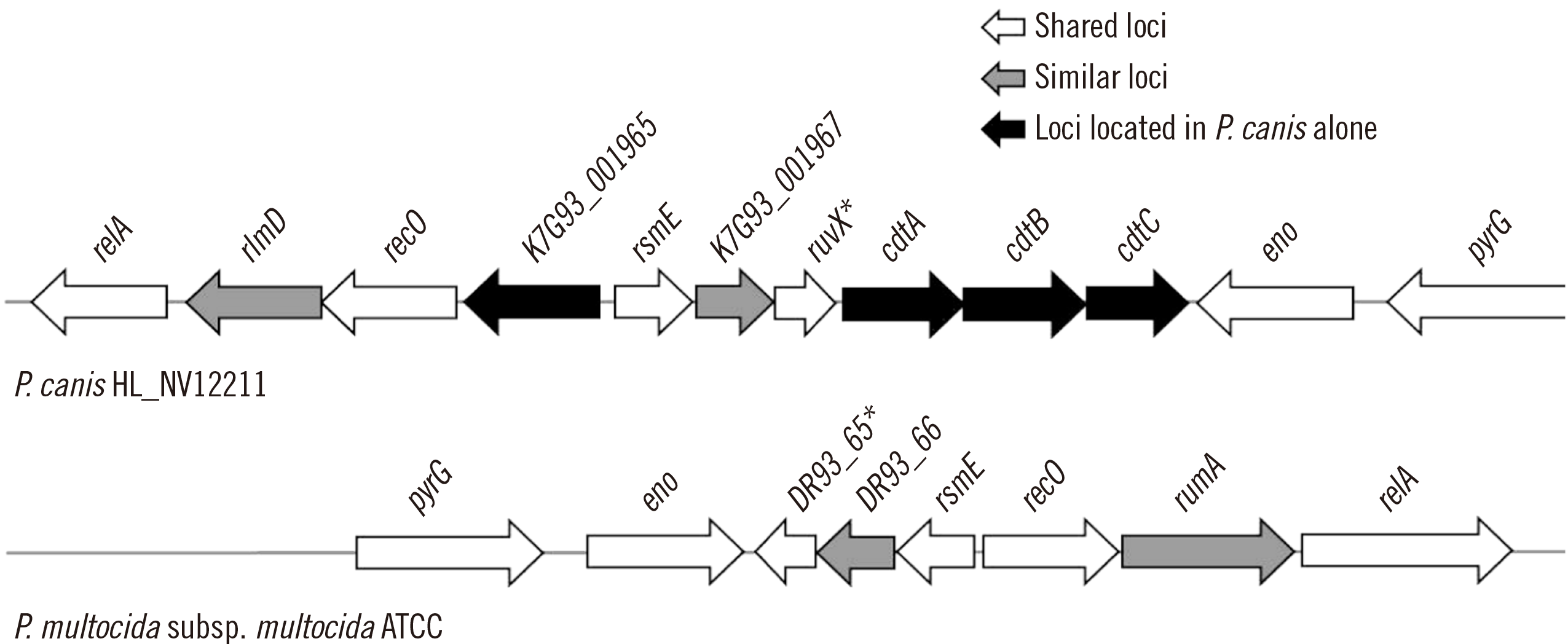
Fig. 3
Gel electrophoresis images of amplified cdtA–cdtB–cdtC products using DNA from clinical isolates. Asterisks indicate the positive control isolate (PA42).
Abbreviations: M, size marker; NC, negative control.
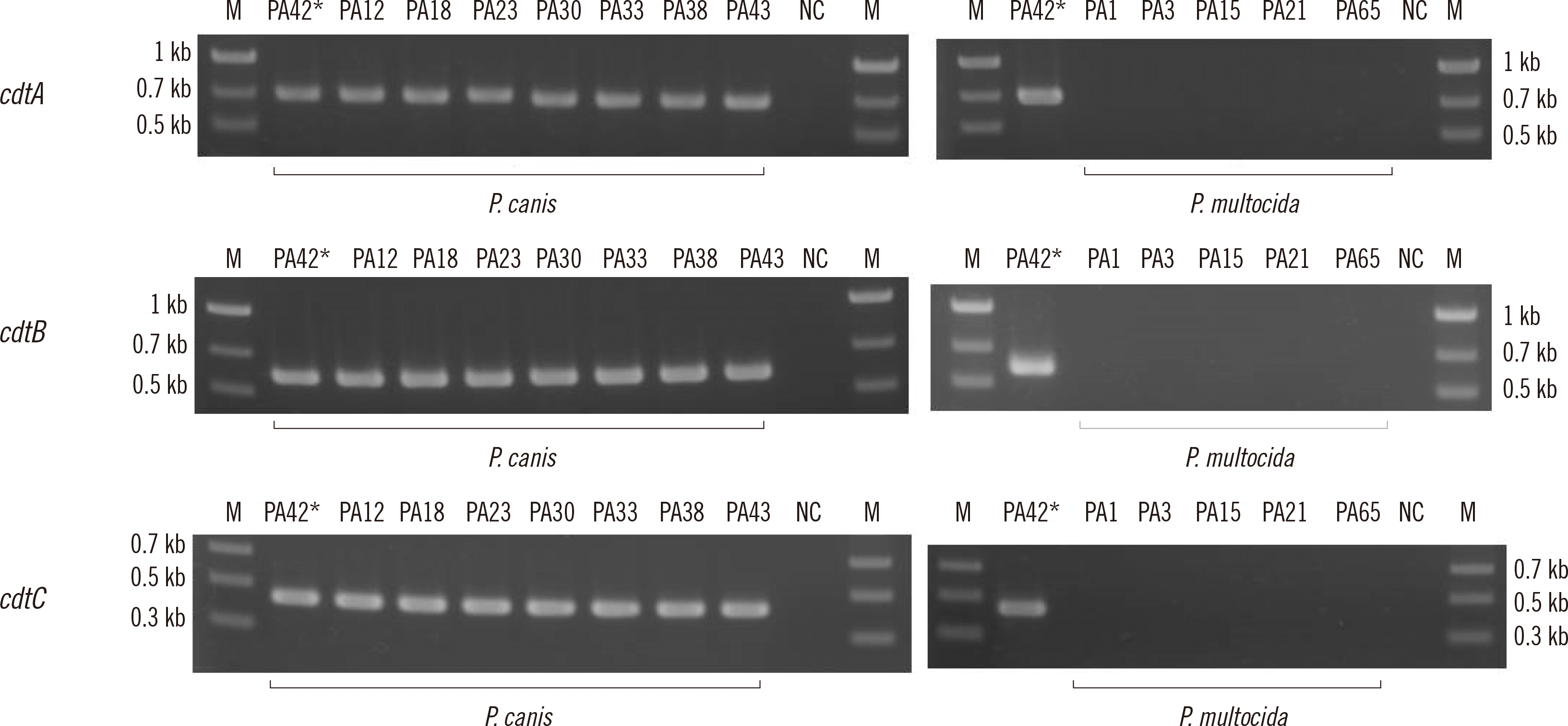
Table 1
Whole-genome sequences of Pasteurella canis and Pasteurella multocida available in the National Center for Biotechnology Information GenBank
| Species or subspecies (sequence type) | Strain | Host | Isolation source | Assembly level | Collection date and location | GenBank accession number |
|---|---|---|---|---|---|---|
| P. canis | NCTC 11621(T) | Dog | Throat | Contig | 1900/1983, unknown | UGTV00000000.1 |
| P. canis | PA42* | Dog | Blood | Contig | 2021, Japan | BPUX00000000.1 |
| P. canis | HL_NV12211† | Dog | Pus | Complete | 2020, Korea | CP085871.1 |
| P. canis | HL_D3081 | Dog | Pus | Complete | 2019, Korea | CP085873.1 |
| P. canis | HL_D1250 | Dog | Throat | Complete | 2018, Korea | CP085791.1 |
| P. canis | PA57* | Human | Pus | Contig | 2021, Japan | BQFX00000000.1 |
| P. canis | NCTC 11650 | Human | Dog bite | Contig | 1900/1984, unknown | UATN00000000.1 |
| P. canis | QBSD | Human | Pus | Contig | 2019, China | WUMP00000000.1 |
| P. canis | HL_1500 | Human | Pus | Complete | 2017, Korea | CP083396.1 |
| P. canis | HL268 | Human | Pus | Complete | 2004, Korea | CP083262.1 |
| P. multocida subsp. multocida (ST3) | NCTC 10322(T)/ATCC 43137(T)† | Porcine | Unknown | Complete | 1900/1962, unknown | LT906458.1 |
| P. multocida subsp. gallicida (ST42) | NCTC 10204(T)/ATCC 51689(T) | Cow | Unknown | Complete | 1900/1960, unknown | LR134298.1 |
| P. multocida subsp. multocida (ST135) | S298D | Dog | Oral swab | Contig | 2016, Greece | PSQH00000000.1 |
| P. multocida subsp. septica (ST30) | KVNON-213 | Cat | Nasal cavity | Complete | 2018, Korea | CP049756.1 |
| P. multocida subsp. gallicida (ST25) | MSP58 | Cat | Clinical sample | Contig | 2019, USA | SJXC00000000.1 |
| P. multocida subsp. septica (ST43) | NCTC 11995(T)/ATCC 51687(T) | Human | Abscess by cat bite | Contig | 1900/1987, France | UGSV00000000.1 |
| P. multocida subsp. multocida (ST131) | NCTC 10382 | Human | Infected finger | Complete | 1964, unknown | LS483473.1 |
| P. multocida subsp. multocida (ST135) | PY81579 | Human | Pus | Contig | 2016, Greece | PSQI00000000.1 |
| P. multocida subsp. septica(new ST; allele 18-21-28-15-48-17-45) | NCTC 11619 | Human | Wound | Complete | 1900/1983, unknown | LR134514.1 |
| P. multocida subsp. septica(new ST; allele 11-21-32-16-18-17-55) | NCTC 11620 | Human | Unknown | Contig | 1900/1983, unknown | UGSW00000000.1 |
| P. multocida subsp. septica(new ST; allele 18-56-37-16-18-18-55) | FDAARGOS_384 | Human | Abscess | Complete | 2015, USA | CP023516.1 |
| P. multocida subsp. septica(new ST; allele 18-56-37-16-18-18-55) | FDAARGOS_385 | Human | Wound | Complete | 2015, USA | CP023972.1 |
| P. multocida subsp. gallicida (ST128) | HuN001 | Human | Unknown | Complete | 2021, China | CP073238.1 |
| P. multocida subsp. septica (ST137) | 161215033201-1 | Human | Right lung | Complete | 2016, the Netherlands | CP026744.1 |
| P. multocida subsp. septica(new ST; allele 20-58-28-15-18-17-24) | FDAARGOS_261 | Human | Blood | Contig | 2014, USA | NBTJ00000000.2 |
| P. multocida subsp. multocida(new ST; allele 16-19-14-6-26-12-11) | SMC1 | Human | Finger bone biopsy | Contig | 2015, Malaysia | LNCO00000000.1 |
Table 2
Oligonucleotide primers for targeted genes and their PCR amplicon sizes
| Target gene (encoding protein) | Primer* | Direction | Sequence (5´→ 3´) (length, k-mer) | Tm (°C)† | Expected amplicon size |
|---|---|---|---|---|---|
| cdtA | Pc_cdtA_F | Forward | TCAGCAGATGTGTAATTGTCCTC (23) | 54 | 693 bp |
| (cytolethal distending toxin A) | Pc_cdtA_R | Reverse | ATCGCAGTCGCATTTAATAGC (21) | 54 | |
| cdtB | Pc_cdtB_F | Forward | TCCAAGAGGCGGGTACTTTG (20) | 54 | 582 bp |
| (cytolethal distending toxin B) | Pc_cdtB_R | Reverse | AACTGGCACCAATACGCTCA (20) | 54 | |
| cdtC | Pc_cdtC_F | Forward | GAGTTATCACCACCTCCACGT (21) | 53 | 433 bp |
| (cytolethal distending toxin C) | Pc_cdtC_R | Reverse | GCGGTACTAAAATTTTACTTGGTCCA (26) | 55 |
Table 3
Prevalence of cytolethal distending toxin (cdt) genes in Pasteurella canis and Pasteurella multocida isolated from different hosts
| Target gene | P. canis isolates (N=30) | P. multocida isolates (N=48) | P* | ||
|---|---|---|---|---|---|
|
|
|
||||
| Dog/cat origin (N=17) | Human origin (N=13) | Dog/cat origin (N=32) | Human origin (N=16) | ||
| cdtA | 17/17 | 13/13 | 0/32 | 0/16 | < 0.01 |
| cdtB | 17/17 | 13/13 | 0/32 | 0/16 | < 0.01 |
| cdtC | 17/17 | 13/13 | 0/32 | 0/16 | < 0.01 |




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download