INTRODUCTION
MATERIALS AND METHODS
1. Cell culture
2. Transfection
3. Animals
4. Paw thickness
5. Behavioral tests
6. Mechanical allodynia
7. Cold allodynia
8. Enzyme-linked immunosorbent assay (ELISA)
9. Western blot assay
10. 3-[4,5-dimethyl-2-thiazolyl]-2,5 diphenyl-2H-tetrazolium bromide (MTT) assay
11. Quantitative real-time polymerase chain reaction (qRT-PCR)
Table 1
12. Immunohistochemical (IHC) analysis
13. Cell proliferation assay
14. Transwell assay
15. Statistical analysis
RESULTS
1. HPSE promotes the malignant phenotype of melanoma cells
 | Fig. 1Heparanase (HPSE) promotes the malignant phenotype of melanoma cells. (A) HPSE expression was determined in B16-F10 and A375 cells by western blot. (B) Cell viability was measured by MTT assay in B16-F10 and A375 cells. (C) The proliferation of melanoma cells was detected by the EdU detection kit (200×, scale bar = 100 μm). (D, E) Transwell assay was used to determine cell migration and invasion (200×, scale bar = 200 μm). *P < 0.05, **P < 0.01 versus control. The error bars indicate mean ± standard deviation. MTT: 3-[4,5-dimethyl-2-thiazolyl]-2,5 diphenyl-2H-tetrazolium bromide, EdU: 5-Ethynyl-20-deoxyuridine. |
2. HPSE promotes tumor growth
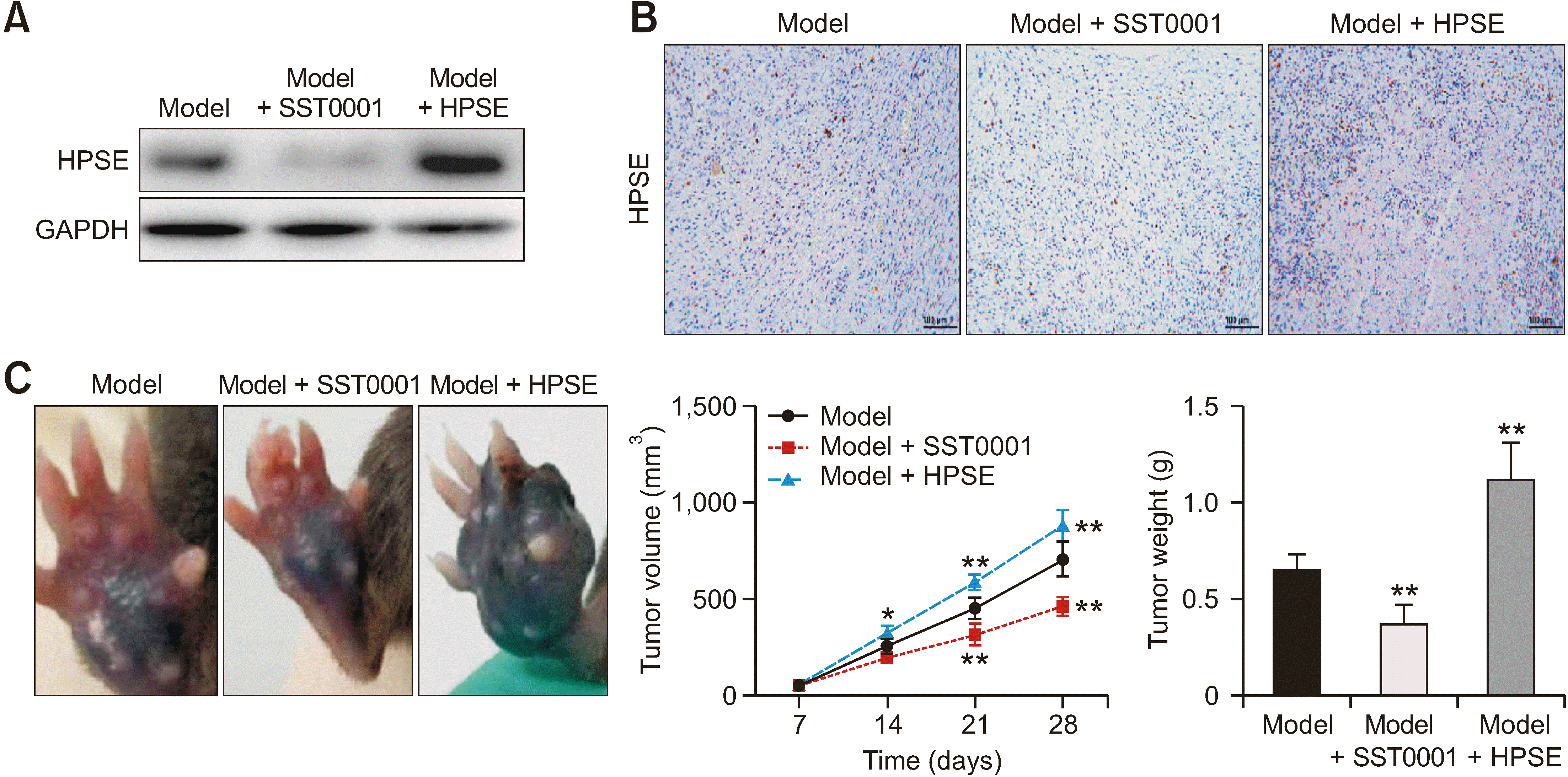 | Fig. 2Heparanase (HPSE) promotes tumor growth. (A) The expression of HPSE was detected by western blot after SST0001 or HPSE treatment. (B) HPSE in tumor tissues was detected by IHC (200×, scale bar = 100 μm). (C) Tumor volume and weight in different groups. *P < 0.05, **P < 0.01 versus model. The error bars indicate mean ± standard deviation. IHC: immunohistochemical. |
3. HPSE promotes cancer pain in mice
 | Fig. 3Heparanase (HPSE) promotes cancer pain in mice. (A) Paw thickness. (B) Mechanical allodynia. (C) Cold response. (D) Spontaneous pain. (E–G) The content of TNF-α, NF-κB, IL-6, IL-1β, and PD-L1 were detected by ELISA assay. (H) PGE2 expression was detected by qRT-PCR. **P < 0.01 versus control. ##P < 0.01 versus model. The error bars indicate mean ± standard deviation. TNF-α: tumor necrosis factor-alpha, NF-κB: nuclear factor-κB, IL: interleukin, PD-L1: programmed death-ligand 1, PGE2: prostaglandin E2, ELISA: enzyme-linked immunosorbent assay, qRT-PCR: quantitative real-time polymerase chain reaction. |
4. HPSE up-regulates NGF and is promoted by NGF feedback
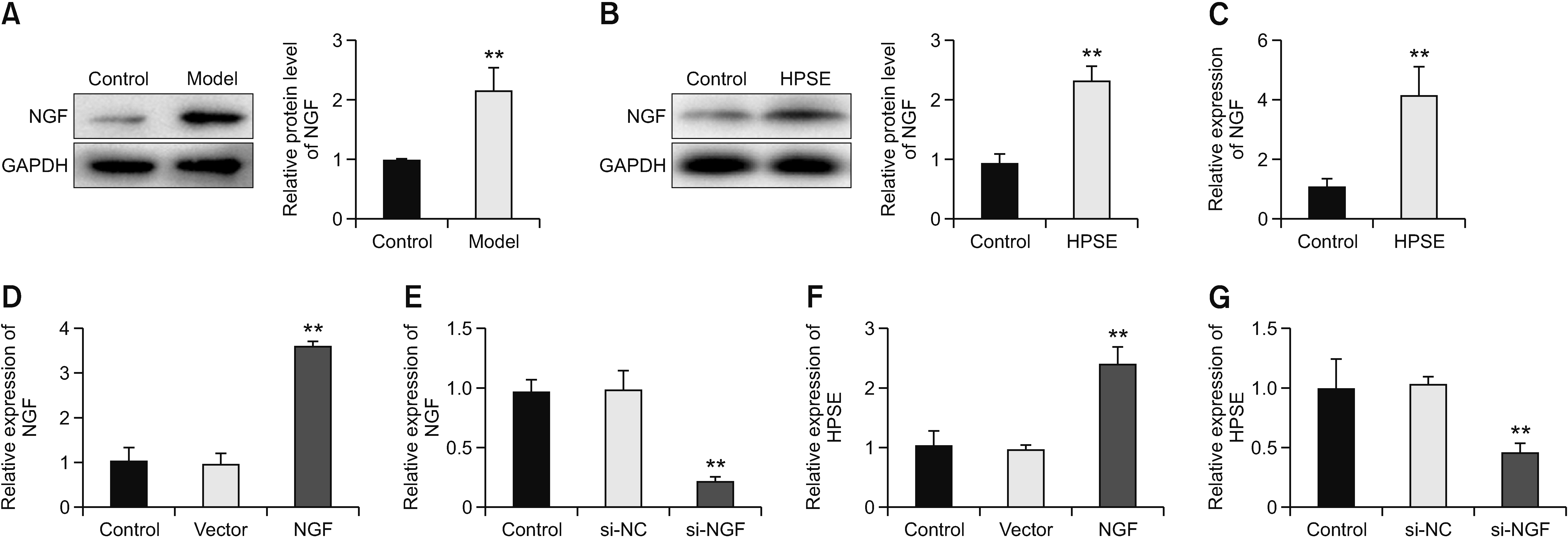 | Fig. 4Heparanase (HPSE) up-regulates nerve growth factor (NGF) and is promoted by NGF feedback. (A) The expression of NGF in the model and control mice was detected by western blot. (B, C) The expression of NGF was detected by western blot and qRT-PCR after HPSE treatment. **P < 0.01 versus control. (D) The expression of NGF was determined by qRT-PCR after NGF treatment. **P < 0.01 versus vector. (E) The expression of NGF was detected by qRT-PCR after NGF silencing. **P < 0.01 versus si-NC. (F) The expression of HPSE was detected by qRT-PCR after NGF treatment. **P < 0.01 versus vector. (G) The expression of HPSE was detected by qRT-PCR after NGF silencing.**P < 0.01 versus si-NC. The error bars indicate mean ± standard deviation. qRT-PCR: quantitative real-time polymerase chain reaction, si-NC: siRNA negative control. |
5. High expression of NGF reverses the inhibition of SST0001 on the malignant phenotype of melanoma cells
 | Fig. 5High expression of nerve growth factor (NGF) reverses the inhibition of SST0001 on the malignant phenotype of melanoma cells. (A) Cell viability was measured by MTT assay in B16-F10 cells. (B) The proliferation of melanoma cells was detected by the EdU detection kit (200×, scale bar = 100 μm). (C) Transwell assay was used to determine cell migration and invasion (200×, scale bar = 200 μm). *P < 0.05, **P < 0.01 versus control. #P < 0.05, ##P < 0.01 versus SST0001. The error bars indicate mean ± standard deviation. MTT: 3-[4,5-dimethyl-2-thiazolyl]-2,5 diphenyl-2H-tetrazolium bromide, EdU: 5-Ethynyl-20-deoxyuridine. |
6. HPSE promotes cancer pain in mice by interacting with SDC1
 | Fig. 6Heparanase (HPSE) promotes cancer pain in mice by interacting with Syndecan-1 (SDC1). (A) The expression of SDC1 was detected by western blot after SST0001 treatment. (B) SDC1 in tumor tissues was detected by IHC (200×, scale bar = 100 μm). (C) The expression of SDC1 was detected by qRT-PCR after the treatment of Ad-SDC1 and/or SST0001. (D) Paw thickness. (E) Mechanical allodynia. (F) Cold response. (G) Spontaneous pain. (H–J) The content of TNF-α, NF-κB, IL-6, IL-1β, and PD-L1 were determined by ELISA assay. (K) PGE2 expression was detected by qRT-PCR. **P < 0.01 versus control. ##P < 0.01 versus model. $P < 0.05, $$P < 0.01 versus Model + SST0001. The error bars indicate mean ± standard deviation. IHC: immunohistochemical, qRT-PCR: quantitative real-time polymerase chain reaction, TNF-α: tumor necrosis factor-alpha, NF-κB: nuclear factor-κB, IL: interleukin, PD-L1: programmed death-ligand 1, PGE2: prostaglandin E2, ELISA: enzyme-linked immunosorbent assay, IOD: integrated optical density. |




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download