Abstract
Paracoccus yeei is rare, opportunistic pathogen which can cause various diseases. Here, we report a case of peritonitis caused by P. yeei, which was successfully treated with cefazolin and gentamicin without catheter removal. In addition, we investigated the presence of several putative virulence genes that showed higher similarity with a previously characterized Korean isolate than with an isolate from the USA.
초록
Paracoccus yeei는 다양한 기회감염을 일으킬 수 있는 드문 균이다. 저자들은 카테터를 제거하지 않고 cefazolin과 gentamicin 투여로 성공적으로 치료된 P. yeei에 의한 복막염 1예를 문헌 고찰과 함께 보고하고자 한다. 더불어 저자들은 추정상의 병독성 유전자(putative virulence gene)의 존재를 조사하였으며 본 증례에서 분리된 균주의 PCR 결과 미국의 P. yeei보다 한국의 P. yeei 균주와 병독성 유전자의 분포가 유사하다는 사실을 밝혀내었다.
Organisms of the genus Paracoccus are common components of the microbiomes of environments such as soil, brines, and biofilters [1, 2]. Paracoccus was formerly categorized as belonging to CDC Group EO-2, which includes aerobic bacteria, with forms ranging from coccoid to short, thick rods, often appearing vacuolated or peripherally stained (O-shaped) [1]. The species P. yeei is unique within the genus because it is associated with human infection [3].
Although P. yeei is rare opportunistic human pathogen, it is capable of causing various human infections including bacteremia [4], keratitis [5], otitis [1], and myocarditis in transplanted hearts [6]. P. yeei has a peculiar characteristic in that it can grow using propylene glycol, a common ingredient in cosmetics, as its sole source of carbon and energy [7].
Here, we describe a case of peritoneal dialysis-related peritonitis caused by P. yeei, which was successfully treated with cefazolin and gentamicin without catheter removal. We investigated several putative virulence determinants and concluded that, to our knowledge, this is the primary peritonitis-causing P. yeei isolate in Korea.
A 39-year-old male with end-stage renal failure due to focal segmental glomerulosclerosis had been on continuous ambulatory peritoneal dialysis since 2012. He was seen at the PD peritoneal dialysis unit on March 21, 2021 reporting cloudy peritoneal fluid in the absence of fever or abdominal pain. Microscopic examination of the peritoneal fluid showed 1,750/μL of white blood cells (WBCs), with 46% of those being neutrophils. Other laboratory tests such as complete blood cell count or C-reactive protein (CRP) were not performed.
A peritoneal fluid sample of approximately 5 mL was inoculated in a BACTEC Plus Aerobic/F and Plus Anaerobic/F broth (Becton Dickinson, Meylan, France) and incubated at 37°C in a BACTEC blood culture system (Becton Dickinson). Bacterial growth was detected in the aerobic bottle after 30 hours of incubation. This was subsequently subcultured onto blood, McConkey, and chocolate agar. After 48 hours of incubation, shiny, brownish white colonies were observed on the blood and chocolate agars (Fig. 1A and 1B). Gram staining of these colonies showed the presence of gram-negative diplococci with a central halo shaped like a doughnut (Fig. 2). This strain (P. yeei CUK22150) was identified as P. yeei with 99.9% confidence using VITEK-MS (BioMérieux, Marcy-l’Etoile, France) and VITEK 2 GN card (BioMérieux, Marcy l’Etoile, France). For confirmation, sequencing of the 16S ribosomal RNA gene was performed using a universal primer pair: 27F (5′-AGAGTTTGATCC TGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGA CTT-3′) [8]. When compared to sequences found in the NCBI database using the BLAST search engine, the sequence showed 100% identity (1329 of 1329 bases) to P. yeei NR_029038.1. An antimicrobial susceptibility test was performed using the disk diffusion method on Muller-Hinton agar. As no interpretative criteria exist for the antimicrobial susceptibility testing of P. yeei, we adopted the CLSI M100-ED31 criteria for the P. aeruginosa disk diffusion breakpoint [9]. Our sample was susceptible to piperacillin/tazobactam, amikacin, cefoxitin, ceftazidime, ceftriaxone, amoxicillin/clavulanate, cefepime, meropenem, imipenem, and ertapenem.
Recently, Lasek et al. [3] characterized the species-specific genes of P. yeei and predicted the putative determinants of virulence. We therefore investigated several putative virulence genes, namely PY32053_00015, PY32053_00016, PY32053_00072, PY32053_00730, PY32053_00902, PY32053_04123, PY32053_04126, PY32053_04669, PY32053_04670, and PY32053_04680, the predicted functions of which are peptide-methionine (S)-S-oxide reductase (MsrA1), peptide-methionine (R)-S-oxide reductase (Msr B), diguanylate cyclase, superoxide dismutase, sugar transferase, urease subunit alpha UreA, urease accessory protein UreD, virB10-like T4SS protein, virB9-like T4SS protein, and lytic murein transglycosylase, respectively (Table 1), using primers manufactured in this study using the Primer3Plus program [10] (Supplementary Table 1). We conducted a PCR analysis, which was positive for PY32053_00015, PY32053_00730, PY32053_00072, PY32053_00902, PY32053_04123, and PY32053_04126 (Fig. 3, Table 1). All products were between 300 and 350 bp in length.
After administration of intraperitoneal cefazolin and gentamicin, the turbidity of the peritoneal fluid gradually cleared and the WBC count was normalized. Although the negative conversion of peritoneal dialysis fluid culture was achieved after 3 days, the patient received intraperitoneal administration of gentamicin for 3 weeks according to the ISPD peritonitis recommendations [11].
The genus Paracoccus consists of over 50 species which share common features such as nonmotility and oxidase positivity [1]. P. yeei, however, is unique because of its capability to infect humans. Lasek et al. [3] investigated the genome composition of P. yeei to gain insight into its mechanisms of pathogenicity and reported 25 putative virulence determinants in the P. yeei CCUG 32053 strain (Sweden). There They were compared with the genomes of two other P. yeei strains; P. yeei TT13 from the forehead skin of a college student from Korea [7] and P. yeei FDAARGOS 252 isolated from urine suprapubic aspirates in the USA (GenBank accession number CP020442.2). Among the 25 putative virulence determinants, six were located on chromosomes and the remaining 19 were located on two extrachromosomal replicons (pYEE2 and pYEE5). Interestingly, while several genes located in chromosomal genomic islands (GIpye 2) were not detected in any of the three strains, the 7 genes located in pYEE2 were detected in all three strains [3]. In addition, the distribution of the 25 putative virulence determinants reported differed between strains.
In this study, we used PCR to identify the presence of virulence genes as analysis of the virulence gene profile is important for infection prognosis and understanding the epidemiological prevalence. We investigated the presence of each of the five determinants in the chromosome and extrachromosomal replicons by PCR and sequencing. The PY32053_00015, PY32053_00072, PY32053_00730, PY32053_00902, PY32053_04123, and PY32053_04125 genes were found to be amplified. This suggests that the strain of P. yeei used in this study contains several virulence genes related to resistance to oxidative stress, formation of biofilm, and adjustment to pH conditions. In addition, P. yeei CUK 22150 showed the highest concordance with P. yeei TT13 (9 determinants), followed by P. yeei CGUG 32053 (6 determinants) and P. yeei FDA-ARGOS_252 (3 determinants), respectively (Table 1) [3, 7, 12].
P. yeei CUK 22150 was susceptible to piperacillin/tazobactam, amikacin, cefoxitin, ceftazidime, ceftriaxone, amoxicillin/clavulanate, cefepime, meropenem, imipenem, and ertapenem. This is in line with a study by Arias et al. [13] who found that P. yeei was susceptible to gentamicin, amikacin, piperacillin/tazobactam, imipenem, and meropenem, although the broth microdilution method and EUCAST breakpoints for Pseudomonas and Acinetobacter spp. were used. Wallet et al. [2] also reported similar antimicrobial susceptibility test results and recommended the intraperitoneal administration of antibiotics for a more concentrated dose leading to higher concentrations at the infected site [2].
We reviewed 5 other cases of peritoneal dialysis-related peritonitis caused by P. yeei (Table 2) and found some common features between among the 6 patients; symptoms were usually mild with an absence of fever and little or no abdominal pain, and treatment outcomes were satisfactory even without dialysis catheter removal [2, 13-16].
Cases like this can often be misinterpreted as caused by coagulase-negative staphylococcus (CoNS), which is the most common cause of peritoneal dialysis-related peritonitis. Misdiagnosis of a P. yeei infection as one caused by CoNS is critical because the treatment of CoNS peritonitis is intraperitoneal cephalosporins or vancomycin for 2 weeks, which is ineffective for that of a P. yeei infection, which should be treated with effective antibiotics for 3 weeks [11]. The introduction of new technologies such as MALDI-TOF MS and 16s rRNA sequencing has made it possible to identify peritonitis-causing pathogens rapidly and accurately.
The 2016 ISPD peritonitis recommendations [11] state that for empiric antibiotic selection, gram-negative organisms should be covered by third-generation cephalosporin or an aminoglycoside. However, after identification of the organisms, while Pseudomonas peritonitis should be treated with 2 antibiotics with different mechanisms of action, non-Pseudomonas gram-negative peritonitis can be treated with other effective antibiotics. Therefore, rapid and accurate identification of causative organisms and subsequent antimicrobial susceptibility are important for proper antibiotic selection and treatment. We present the first case of P. yeei-caused peritoneal dialysis-related peritonitis in Korea, which was successfully managed with the administration of intraperitoneal gentamicin and cefazolin.
REFERENCES
1. Daneshvar MI, Hollis DG, Weyant RS, Steigerwalt AG, Whitney AM, Douglas MP, et al. 2003; Paracoccus yeeii sp. nov. (formerly CDC group EO-2), a novel bacterial species associated with human infection. J Clin Microbiol. 41:1289–94. DOI: 10.1128/JCM.41.3.1289-1294.2003. PMID: 12624070. PMCID: PMC150304.
2. Wallet F, Blondiaux N, Foy CL, Loïez C, Armand S, Pagniez D, et al. 2010; Paracoccus yeei: a new unusual opportunistic bacterium in ambulatory peritoneal dialysis. Int J Infect Dis. 14:e173–4. DOI: 10.1016/j.ijid.2009.03.030. PMID: 19556157.
3. Lasek R, Szuplewska M, Mitura M, Decewicz P, Chmielowska C, Pawłot A, et al. 2018; Genome structure of the opportunistic pathogen Paracoccus yeei (Alphaproteobacteria) and identification of putative virulence factors. Front Microbiol. 9:2553. DOI: 10.3389/fmicb.2018.02553. PMID: 30410477. PMCID: PMC6209633.
4. Aliste-Fernández M, Sanfeliu-Sala I, Sánchez-Delgado J. 2020; Bacteremia caused by Paracoccus yeei in patient with compensated cirrhosis of the liver. Enferm Infecc Microbiol Clin (Engl Ed). 38:451–2. DOI: 10.1016/j.eimc.2020.01.013. PMID: 32127236.
5. Courjaret JC, Drancourt M, Hoffart L. 2014; Paracoccus yeei keratitis in a contact lens wearer. Eye Contact Lens. 40:e21–2. DOI: 10.1097/ICL.0b013e31829e8fc7. PMID: 24045834.
6. Schweiger M, Stiegler P, Scarpatetti M, Wasler A, Sereinigg M, Prenner G, et al. 2011; Case of Paracoccus yeei infection documented in a transplanted heart. Transpl Infect Dis. 13:200–3. DOI: 10.1111/j.1399-3062.2010.00571.x. PMID: 20854281.
7. Lim JY, Hwang I, Ganzorig M, Pokhriyal S, Singh R, Lee K. 2018; Complete genome sequence of Paracoccus yeei TT13, isolated from human skin. Genome Announc. 6:e01514–7. DOI: 10.1128/genomeA.01514-17. PMID: 29348361. PMCID: PMC5773746.
8. Clinical, Laboratory Standards Institute. 2018. Interpretive criteria for identification of bacteria and fungi by targeted DNA sequencing. 2nd ed. CLSI guideline MM18. Wayne, PA: Clinical and Laboratory Standards Institute.
9. Clinical, Laboratory Standards Institute. 2021. Performance standards for antimicrobial susceptibility testing. 31st ed. CLSI supplement M100. Wayne PA: Clinical and Laboratory Standards Institute.
10. Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, et al. 2012; Primer3-new capabilities and interfaces. Nucleic Acids Res. 40:e115. DOI: 10.1093/nar/gks596. PMID: 22730293. PMCID: PMC3424584.
11. Li PK, Szeto CC, Piraino B, de Arteaga J, Fan S, Figueiredo AE, et al. 2016; ISPD Peritonitis Recommendations: 2016 Update on Prevention and Treatment. Perit Dial Int. 36:481–508. DOI: 10.3747/pdi.2016.00078. PMID: 27282851. PMCID: PMC5033625.
12. Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. 2016; GenBank. Nucleic Acids Res. 44:D67–72. DOI: 10.1093/nar/gkv1276. PMID: 26590407. PMCID: PMC4702903.
13. Arias MA, Clark J. 2019; Paracoccus yeei as a cause of peritoneal dialysis peritonitis in the United Kingdom. IDCases. 15:e00486. DOI: 10.1016/j.idcr.2019.e00486. PMID: 30701158. PMCID: PMC6348235.
14. Sastre A, González-Arregoces J, Romainoik I, Mariño S, Lucas C, Monfá E, et al. 2016; Paracoccus yeei peritonitis in peritoneal dialysis. Nefrologia. 36:445–6. DOI: 10.1016/j.nefro.2016.02.009. PMID: 27039709.
15. Palamuthusingam D, Tan KS. 2014; The first case of Paracoccus yeeii species infection in Australia causing peritonitis in an APD patient. Nephrology (Carlton). 19:116. DOI: 10.1111/nep.12167. PMID: 24428219.
16. Nakayama T, Morimoto K, Uchiyama K. 2021; Peritoneal dialysis-related peritonitis caused by Paracoccus yeei. Ther Apher Dial. 25:715–7. DOI: 10.1111/1744-9987.13604. PMID: 33135289.
Fig. 1
(A) Colonies of Paracoccus yeei on a blood agar plate after 24 hours of incubation in a 37°C incubator with 5% CO2. (B) Colonies of P. yeei on a blood agar plate after 48 hours of incubation. Note the shiny brownish white colonies.
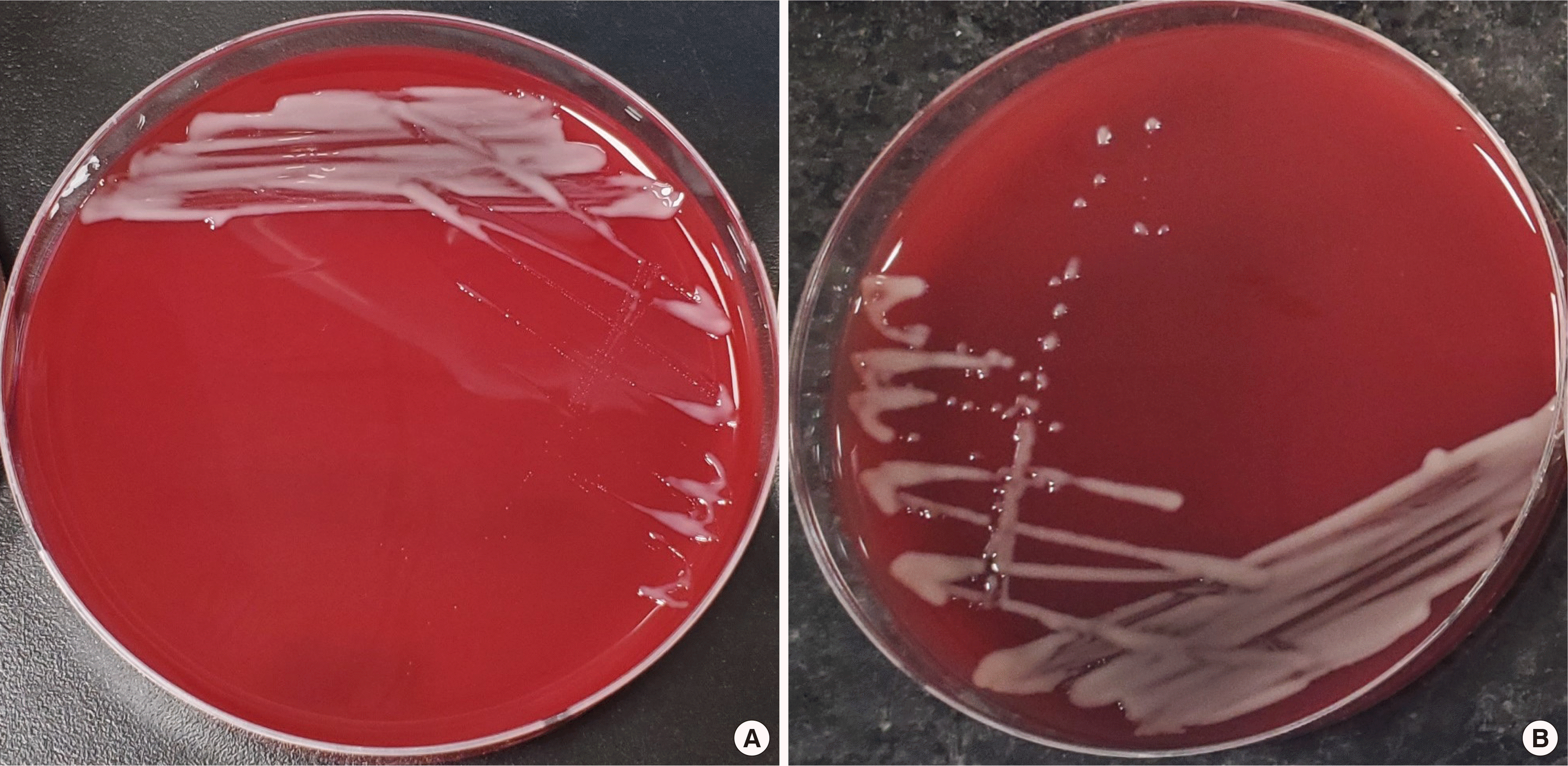
Fig. 2
Microscopic description of Paracoccus yeei with Gram stain. Note the gram-negative diplococci with peripheral staining (O-shaped).
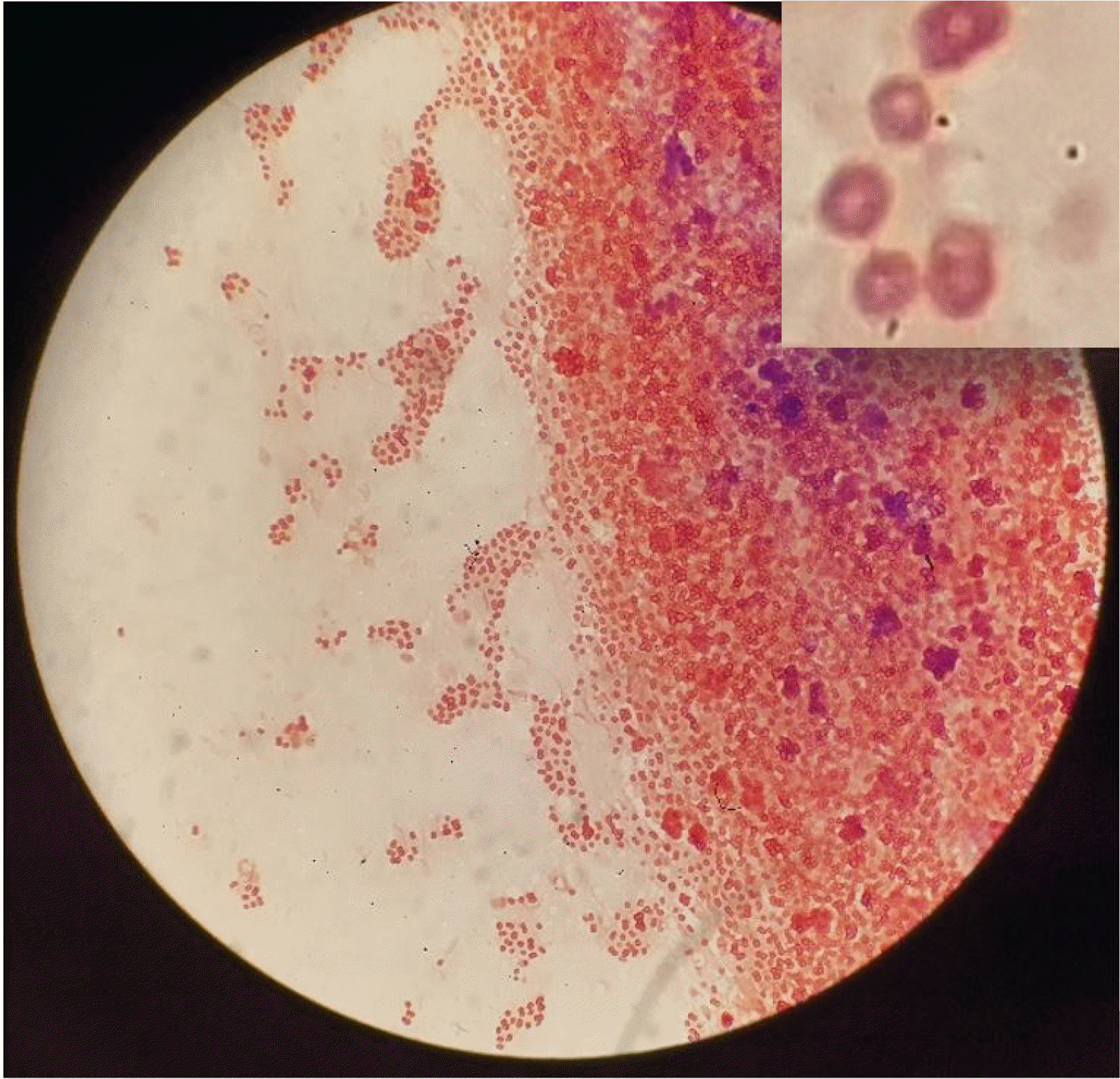
Fig. 3
1.5% agarose gel electrophoresis results from the amplification of putative virulence genes. Lane L contains the ladder and lane N contains the negative control with primer and no Paracoccus yeei DNA. Lane P contains the KPC gene product with a KPC-specific primer (873 bp). Lanes 1–10 contain virulence genes (PY32053_00730, PY32053_00015, PY32053_00016, PY32053_00072, PY32053_00902, PY32053_04123, PY32053_04125, PY32053_04669, PY32053_04670, PY32053_04680).
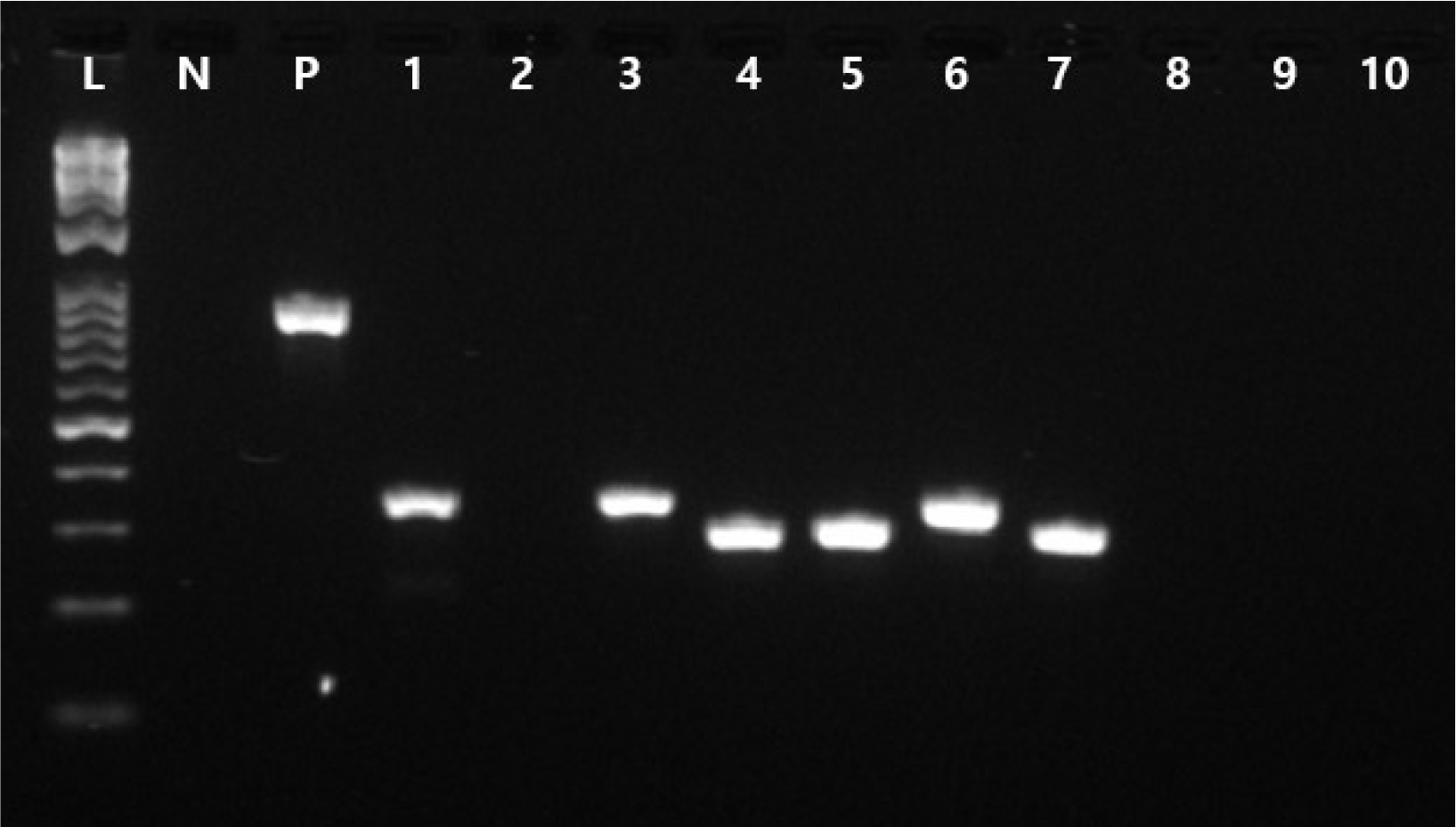
Table 1
Presence or absence of putative virulence genes of four Paracoccus yeei isolates
| Gene location | Gene locus name | Predicted function of gene product | Gene presence | |||
|---|---|---|---|---|---|---|
|
|
||||||
| P. yeei CCUG 32053 [3] | P. yeei TT13 (skin sample, Korea) [7] | P. yeei FDAARGOS_252 (urine sample, USA, GenBank accession number CP020442) [12] | P. yeei in this case (P. yeei CUK 21150) | |||
| chr (GIpye 2) | PY32053_00015 | Peptide-methionine (S)-S-oxide reductase (MsrA1) | + | + | - | + |
| PY32053_00016 | Peptide-methionine (R)-S-oxide reductase (MsrB) | + | + | +, 55% similarity | - | |
| chr (GIpye 2) | PY32053_00072 | Diguanylate cyclase | + | + | - | + |
| chr | PY32053_00730 | Superoxide dismutase | + | + | + | + |
| chr | PY32053_00902 | Sugar transferase | + | + | + | + |
| pYEE2 | PY32053_04123 | Urease subunit alpha UreA | + | + | + | + |
| PY32053_04125 | Urease accessory protein UreD | + | + | - | + | |
| pYEE5 | PY32053_04669 | VirB10-like T4SS protein | + | - | + | - |
| PY32053_04670 | VirB9-like T4SS protein | + | - | + | - | |
| PY32053_04680 | Lytic murein transglycosylase | + | - | + | - | |
Table 2
Description of case reports of Paracoccus yeei peritonitis.
| Reference | Age/Sex | Underlying disease | Fever | Abdominal pain | Antibiotics | Outcome |
|---|---|---|---|---|---|---|
| [2] | 25/M | IgA nephropathy | No | Yes | Piperacillin cephalothin | Recovery |
| [13] | 81/M | Diabetic kidney disease | No | No | Vancomycin gentamicin | Recovery |
| [14] | 46/F | Polycystic kidney disease | No | Yes | Vancomycin ceftazidime | Recovery |
| [15] | 72/M | Diabetic kidney disease | No | Yes | Vancomycin gentamicin | Recovery |
| [16] | 51/M | Diabetic kidney disease | No | No | Vancomycin ceftriaxone | Recovery |
| Our case | 39/M | Focal segmental glomerulosclerosis | No | No | Cefazolin gentamicin | Recovery |




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download