1. Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010; 127:2893–917.

2. Labianca R, Beretta GD, Kildani B, Milesi L, Merlin F, Mosconi S, et al. Colon cancer. Crit Rev Oncol Hematol. 2010; 74:106–33.

3. Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014; 383:1490–502.

4. Jalili-Nik M, Soltani A, Moussavi S, Ghayour-Mobarhan M, Ferns GA, Hassanian SM, et al. Current status and future prospective of Curcumin as a potential therapeutic agent in the treatment of colorectal cancer. J Cell Physiol. 2018; 233:6337–45.

5. Kang YJ, Jo JO, Ock MS, Chang HK, Lee SH, Ahn BK, et al. Thymosin β4 was upregulated in recurred colorectal cancers. J Clin Pathol. 2014; 67:188–90.

6. Chen L, Dahlstrom JE, Chandra A, Board P, Rangasamy D. Prognostic value of LINE-1 retrotransposon expression and its subcellular localization in breast cancer. Breast Cancer Res Treat. 2012; 136:129–42.

7. Hur K, Cejas P, Feliu J, Moreno-Rubio J, Burgos E, Boland CR, et al. Hypomethylation of long interspersed nuclear element-1 (LINE-1) leads to activation of proto-oncogenes in human colorectal cancer metastasis. Gut. 2014; 63:635–46.

8. Mima K, Nowak JA, Qian ZR, Cao Y, Song M, Masugi Y, et al. Tumor LINE-1 methylation level and colorectal cancer location in relation to patient survival. Oncotarget. 2016; 7:55098–109.

9. Speek M. Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol Cell Biol. 2001; 21:1973–85.

10. Nigumann P, Redik K, Matlik K, Speek M. Many human genes are transcribed from the antisense promoter of L1 retrotransposon. Genomics. 2002; 79:628–34.

11. Khalid M, Bojang P Jr, Hassanin AA, Bowers EC, Reyes-Reyes EM, Ramos IN, et al. Line-1: implications in the etiology of cancer, clinical applications, and pharmacologic targets. Mutat Res Rev Mutat Res. 2018; 778:51–60.

12. Martin SL. Ribonucleoprotein particles with LINE-1 RNA in mouse embryonal carcinoma cells. Mol Cell Biol. 1991; 11:4804–7.

13. Holmes SE, Singer MF, Swergold GD. Studies on p40, the leucine zipper motif-containing protein encoded by the first open reading frame of an active human LINE-1 transposable element. J Biol Chem. 1992; 267:19765–8.

14. Hohjoh H, Singer MF. Sequence-specific single-strand RNA binding protein encoded by the human LINE-1 retrotransposon. EMBO J. 1997; 16:6034–43.

15. Hohjoh H, Singer MF. Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. EMBO J. 1996; 15:630–9.

16. Khazina E, Weichenrieder O. Non-LTR retrotransposons encode noncanonical RRM domains in their first open reading frame. Proc Natl Acad Sci U S A. 2009; 106:731–6.

17. Feng Q, Moran JV, Kazazian HH Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996; 87:905–16.

18. Wolff EM, Byun HM, Han HF, Sharma S, Nichols PW, Siegmund KD, et al. Hypomethylation of a LINE-1 promoter activates an alternate transcript of the MET oncogene in bladders with cancer. PLoS Genet. 2010; 6:e1000917.

19. Mathias SL, Scott AF, Kazazian HH Jr, Boeke JD, Gabriel A. Reverse transcriptase encoded by a human transposable element. Science. 1991; 254:1808–10.

20. Dombroski BA, Feng Q, Mathias SL, Sassaman DM, Scott AF, Kazazian HH Jr, et al. An in vivo assay for the reverse transcriptase of human retrotransposon L1 in Saccharomyces cerevisiae. Mol Cell Biol. 1994; 14:4485–92.

21. Drongitis D, Rainone S, Piscopo M, Viggiano E, Viggiano A, De Luca B, et al. Epigenetics and cortical spreading depression: changes of DNA methylation level at retrotransposon sequences. Mol Biol Rep. 2016; 43:755–60.

22. Burns KH. Transposable elements in cancer. Nat Rev Cancer. 2017; 17:415–24.

23. Rodic N, Sharma R, Sharma R, Zampella J, Dai L, Taylor MS, et al. Long interspersed element-1 protein expression is a hallmark of many human cancers. Am J Pathol. 2014; 184:1280–6.

24. Swets M, Zaalberg A, Boot A, van Wezel T, Frouws MA, Bastiaannet E, et al. Tumor LINE-1 methylation level in association with survival of patients with stage II colon cancer. Int J Mol Sci. 2016; 18:36.

25. Miglio U, Berrino E, Panero M, Ferrero G, Coscujuela Tarrero L, Miano V, et al. The expression of LINE1-MET chimeric transcript identifies a subgroup of aggressive breast cancers. Int J Cancer. 2018; 143:2838–48.
26. Park M, Dean M, Kaul K, Braun MJ, Gonda MA, Vande Woude G. Sequence of MET protooncogene cDNA has features characteristic of the tyrosine kinase family of growth-factor receptors. Proc Natl Acad Sci U S A. 1987; 84:6379–83.

27. Birchmeier C, Birchmeier W, Gherardi E, Vande Woude GF. Met, metastasis, motility and more. Nat Rev Mol Cell Biol. 2003; 4:915–25.

28. Ma PC, Maulik G, Christensen J, Salgia R. c-Met: structure, functions and potential for therapeutic inhibition. Cancer Metastasis Rev. 2003; 22:309–25.
29. Kammula US, Kuntz EJ, Francone TD, Zeng Z, Shia J, Landmann RG, et al. Molecular co-expression of the c-Met oncogene and hepatocyte growth factor in primary colon cancer predicts tumor stage and clinical outcome. Cancer Lett. 2007; 248:219–28.

30. Boccaccio C, Comoglio PM. MET, a driver of invasive growth and cancer clonal evolution under therapeutic pressure. Curr Opin Cell Biol. 2014; 31:98–105.

31. Chen T, Cai SL, Li J, Qi ZP, Li XQ, Ye LC, et al. Mecp2-mediated epigenetic silencing of miR-137 contributes to colorectal adenoma-carcinoma sequence and tumor progression via relieving the suppression of c-Met. Sci Rep. 2017; 7:44543.

32. Scheri KC, Leonetti E, Laino L, Gigantino V, Gesualdi L, Grammatico P, et al. c-MET receptor as potential biomarker and target molecule for malignant testicular germ cell tumors. Oncotarget. 2018; 9:31842–60.

33. Zhang J, Babic A. Regulation of the MET oncogene: molecular mechanisms. Carcinogenesis. 2016; 37:345–55.
34. Di Renzo MF, Olivero M, Giacomini A, Porte H, Chastre E, Mirossay L, et al. Overexpression and amplification of the met/HGF receptor gene during the progression of colorectal cancer. Clin Cancer Res. 1995; 1:147–54.
35. Ma PC, Tretiakova MS, MacKinnon AC, Ramnath N, Johnson C, Dietrich S, et al. Expression and mutational analysis of MET in human solid cancers. Genes Chromosomes Cancer. 2008; 47:1025–37.
36. Zeng ZS, Weiser MR, Kuntz E, Chen CT, Khan SA, Forslund A, et al. c-Met gene amplification is associated with advanced stage colorectal cancer and liver metastases. Cancer Lett. 2008; 265:258–69.

37. Boromand N, Hasanzadeh M, ShahidSales S, Farazestanian M, Gharib M, Fiuji H, et al. Clinical and prognostic value of the C-Met/HGF signaling pathway in cervical cancer. J Cell Physiol. 2018; 233:4490–6.

38. Luo Y, Ouyang J, Zhou D, Zhong S, Wen M, Ou W, et al. Long noncoding RNA GAPLINC promotes cells migration and invasion in colorectal cancer cell by regulating miR-34a/c-MET signal pathway. Dig Dis Sci. 2018; 63:890–9.

39. Zhu C, Utsunomiya T, Ikemoto T, Yamada S, Morine Y, Imura S, et al. Hypomethylation of long interspersed nuclear element-1 (LINE-1) is associated with poor prognosis via activation of c-MET in hepatocellular carcinoma. Ann Surg Oncol. 2014; 21 Suppl 4:S729–35.

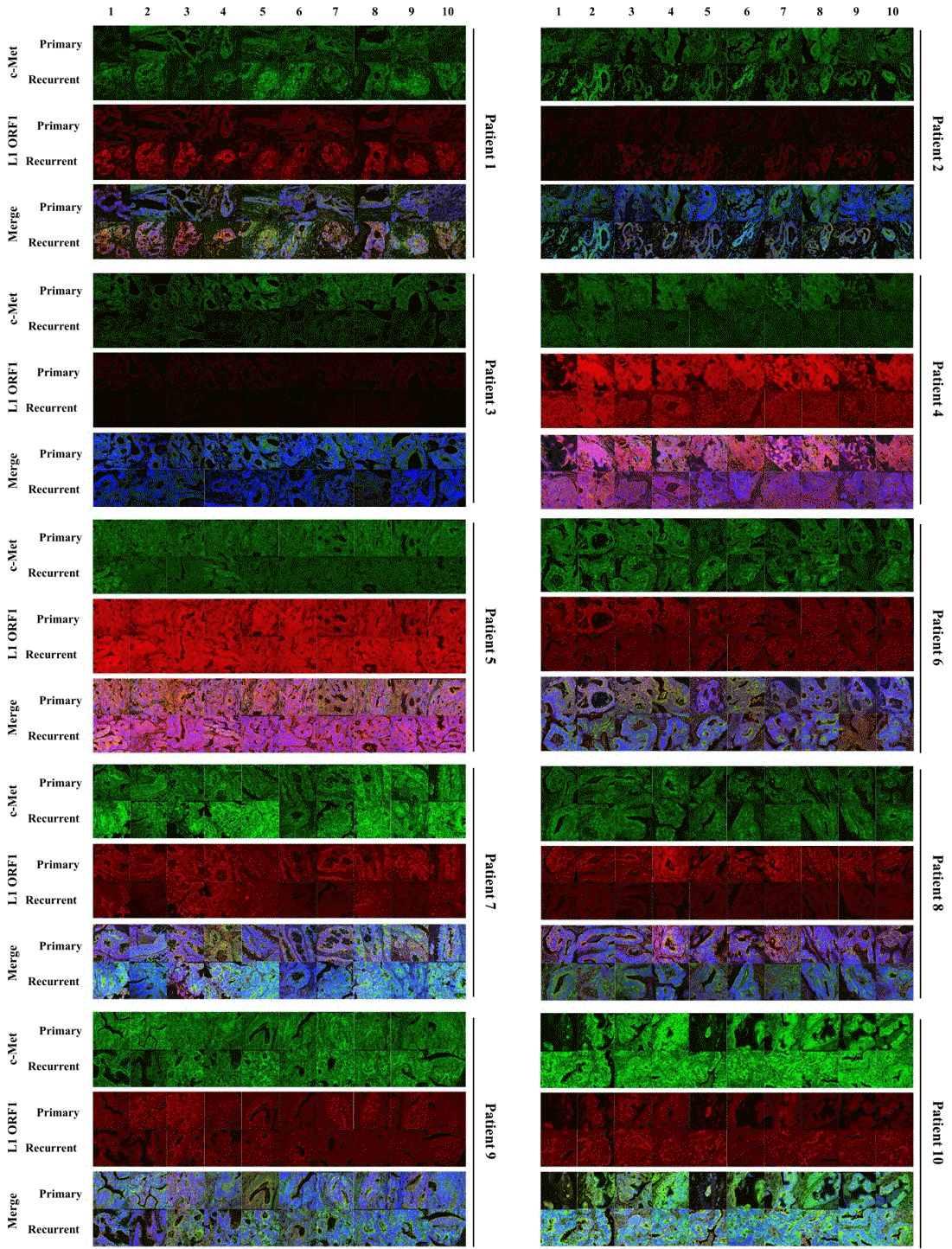
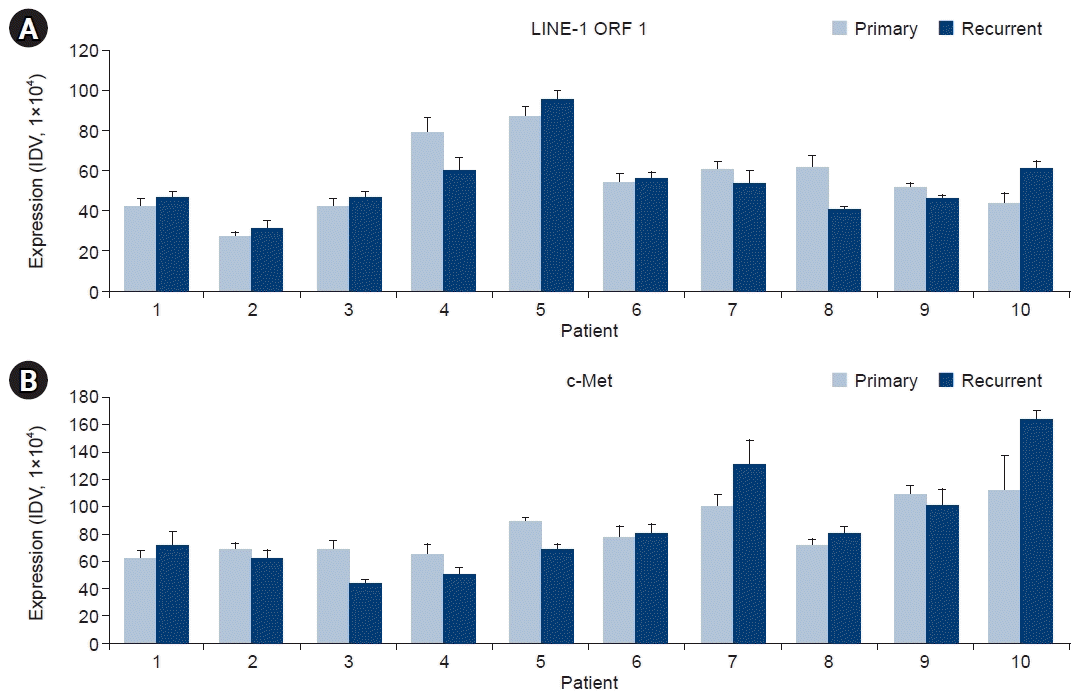
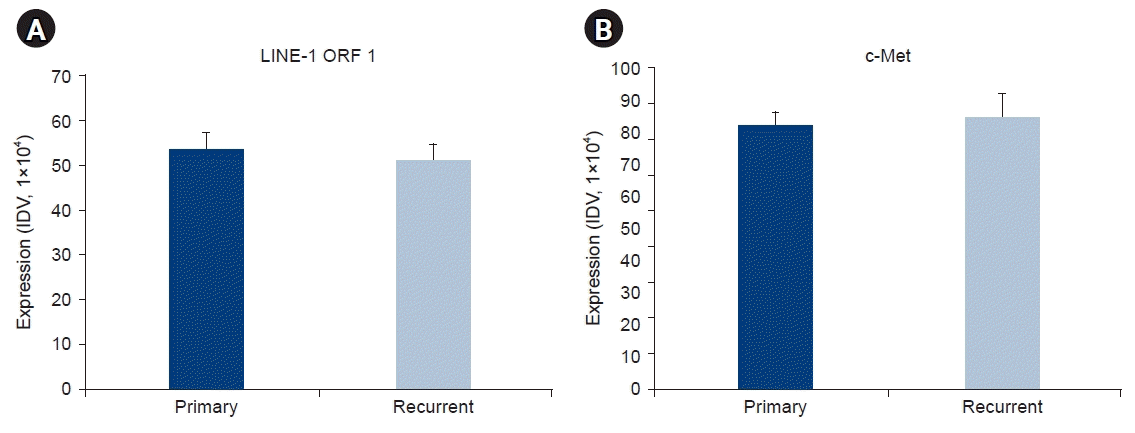
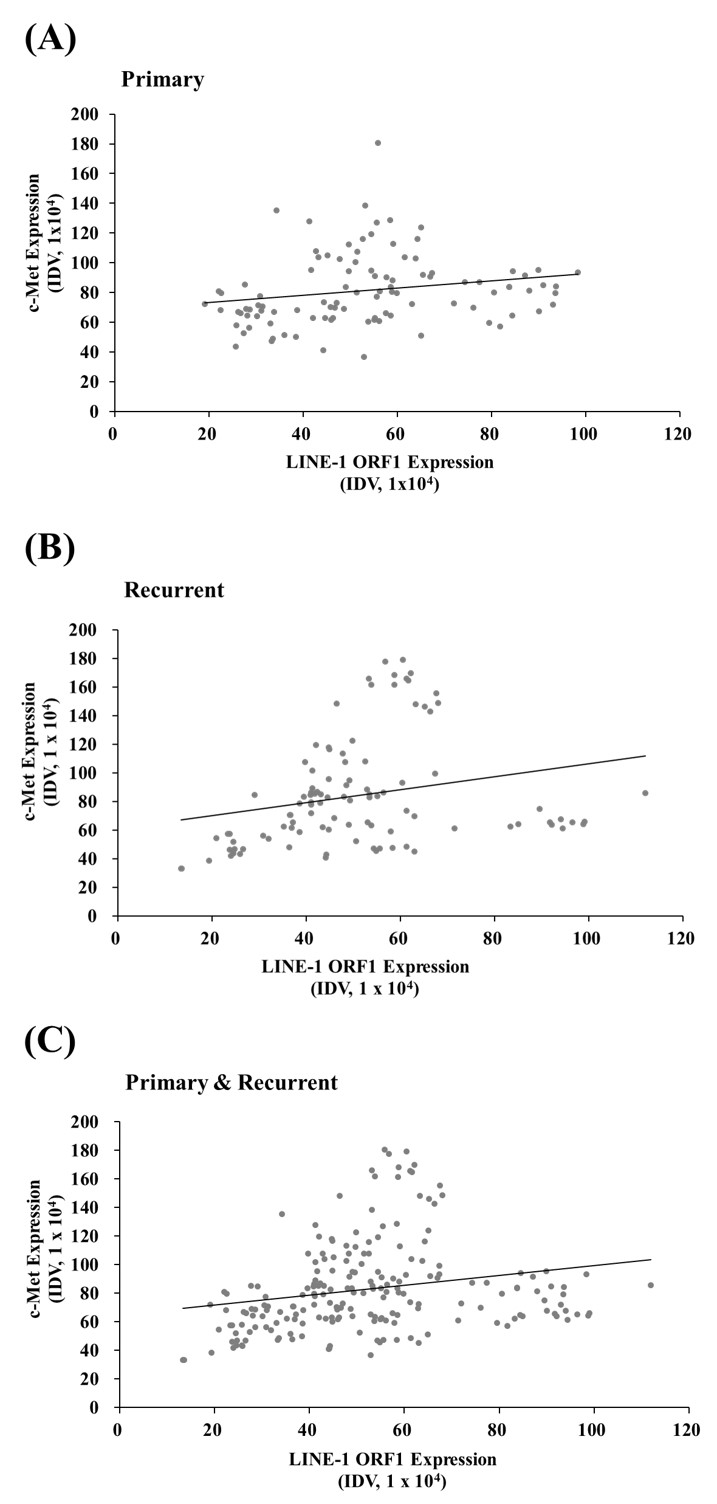




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download