Abstract
Purpose
The prognostic value of vitamin D receptor gene (VDR) expression in breast cancer development is unclear. Here, we aimed to investigate whether VDR expression can be used as a prognostic indicator of breast cancer.
Methods
We used various public bioinformatics platforms: Oncomine, GEPIA, UALCAN, Kaplan-Meier plotter, UCSC XENA, bc-GenExMiner, WebGestalt, and STRING database.
Results
We found that VDR was upregulated in breast cancer in comparison to normal tissues. Overexpression of VDR was significantly associated with worse overall survival in breast cancer. The expression of VDR was related to age, TNM stages, estrogen receptor status, progesterone receptor status, human epidermal growth factor receptor 2 status, basal-like (PAM 50) status, triple-negative breast cancer (TNBC) status, and basal-like (PAM 50) & TNBC status (P < 0.05). Increased VDR expression in breast cancer was significantly associated with older age. The 5 hub genes for VDR were NCOA1, EP300, CREBBP, and RXRA.
Breast cancer is the most common cancer among women worldwide and it is the main leading cause of cancer death in women [1]. Age, gender, benign breast tumors, early menopause, late menarche, hormone therapy, chest radiation exposure, alcohol consumption, diethylstilbestrol use, genetic risk factors, postmenopausal obesity, first pregnancy after the age of 30 years, not breastfeeding, and environmental risk factors are the risk factors for breast cancer [2]. Variations in the degree of gene expression, epigenetic changes, and polymorphisms all have a role in the genetics of breast cancer (DNA sequence 3 alterations).
A growing body of epidemiological evidence suggests an inverse association between vitamin D levels and breast cancer risk, yet the results are still mixed [34]. Vitamin D receptor (VDR) is a ligand-dependent transcription factor in the nuclear receptor superfamily [5]. When it binds to its ligand, calcitriol (1α, 1,25 dihydroxyvitamin D), VDR translocates into the nucleus and binds to the vitamin D response element, activating the transcription of numerous target genes [5] involved in a myriad of cellular functions and processes, and impaired vitamin D activities have been widely implicated in human cancer [67]. In 1979, VDR was first identified in a breast cancer cell line, and was later identified in many breast cancer cell lines and the majority of human breast tumor tissues examined [8]. VDR knockout mice have higher rates of preneoplastic mammary lesions than wild-type mice [8], and treatment with a vitamin D analogue prevents the development of carcinogen-induced mammary tumors [9] by inhibiting cellular proliferation, promoting differentiation, and inducing apoptosis [1011].
Herein, we first assessed the expression pattern of VDR in human breast cancer by bioinformatics analysis. Then, we analyzed the prognostic significance of VDR expression in breast cancer. Finally, we explored several potential mechanisms of VDR action in breast cancer by analysis of co-expressed genes and assessment of its correlation with parental genes. These findings improve our understanding of VDR ’s roles and mechanisms in human breast cancer.
The authors used various public bioinformatics database platforms as follows. The study was approved by the Institutional Review Board committee of Konkuk University Medical Center (No. 20220902).
The Oncomine database (https://www.oncomine.org/resource/login.html) is a web-based data mining platform that incorporates 264 independent datasets and aims to collect, standardize, analyze, and deliver transcriptomic cancer data for biomedical research [12]. An online tumor microarray database was used to determine the expression levels of VDR in breast cancer. The expression fold change of VDR in clinical cancer specimens compared to normal controls was obtained as the following parameters: P-value of 1e-4, fold change of 2, and gene ranking in the top 10%. The co-expression profile of the VDR gene in breast cancer was assessed and displayed as a heat map.
The GEPIA (Gene Expression Profiling Interactive Analysis; http://gepia.cancer-pku.cn/detail.php) is a web server for analyzing the RNA sequencing expression data of 9,736 tumors and 8,587 normal samples from the TCGA (The Cancer Genome Atlas) and the GTEx (Genotype-Tissue Expression) projects, using a standard processing pipeline. The GEPIA-provided box plot tool was used for tumor/normal differential expression analysis of VDR in different cancers [13].
UALCAN (http://ualcan.path.uab.edu/) is an effective website for online analysis and mining of cancer data [14]. It is mainly based on the analysis of relevant cancer data in the TCGA database. It can help us to perform biomarker identification, expression profiling, survival analysis, etc., of related genes. We can query other databases for related information through the links provided.
The Kaplan-Meier plotter (https://kmplot.com/analysis/) is a web-based tool used to analyze the impact of various genes on the survival of cancer patients [15]. We used this tool to analyze the correlations between the expression of VDR and the survival of patients with breast cancers.
bc-GenExMiner (breast cancer Gene-Expression Miner) ver. 4.2 (http://bcgenex.centregauducheau.fr/BC-GEM/GEM-Accueil.php?js=1) was used to analyze the relationship between LEP expression and clinicopathological parameters in breast cancer patients [16].
From National Center for Biotechnology Information (NCBI)-GEO (Gene Expression Omnibus; https://www.ncbi.nlm.nih.gov/gds/), 2 gene expression datasets (GSE27155, GSE35570) were downloaded and analyzed on the GPL570 platform (Affymetrix Human Genome U133 Plus 2.0 array, Thermo Fisher Scientific, Inc., Waltham, MA, USA).
To characterize the functional roles of the differentially expressed genes (DEGs), the functional enrichment analysis web tool WebGestalt (WEB-based Gene SeT AnaLysis Toolkit; http://www.webgestalt.org/) was used for gene ontology (GO) enrichment analysis of molecular function (MF), biological process (BP), and cellular component (CC) with a cutoff of P < 0.05. WebGestalt was also used to explore Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways to understand the signaling pathways in which DEGs are involved [17].
STRING database (https://string-db.org/) [18] is an online tool that provides information on hypothetical gene function, interrogates gene lists, and ranks genes based on functional evaluations. It contains a large set of functional association data, including protein and gene interactions, pathways, and co-expression data. We applied the STRING database analysis tool to predict protein-protein interactions (PPIs) using VDR as a query. The prediction output is in the form of a network that shows the relationships between genes in the list, where nodes symbolize genes and links represent networks. Cytoscape (ver. 3.6.1 [18]) software (http://www.cytoscape.org/) was used to visualize and analyze the PPI network. Hub genes in the network were subsequently identified using the Cytoscape plugin cytoHubba [19].
The heat map and correlation between VDR in the same patient cohort were analyzed by data mining in TCGA-breast cancer using the UCSC (the University of California, Santa Cruz) Xena browser (https://xena.ucsc.edu/) [20]. Subsequently, the methylation status of VDR was also examined using the UCSC Xena browser.
Expression data were extracted from the Oncomine, cBioPortal (https://www.cbioportal.org), and GEPIA databases. The P-values of <0.05 were considered significant. Methylation data were extracted from TCGA UCSC Xena browser, and the box plot was retrieved from UALCAN web. The unpaired t-test was used to determine the P-values, where P < 0.05 was considered significant (*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001). Survival curves were extracted from the Kaplan-Meier scanner, Kaplan-Meier plotter. All survival results are displayed with P-values obtained using the log-rank test. Log-rank P-values < 0.05 were considered significant.
The Oncomine database was used to compare the messenger RNA (mRNA) levels of VDR in breast cancer tissues and normal tissues. The results showed that in comparison to their expression levels in normal tissues, VDR was overexpressed in breast cancer tissues (P < 0.01) (Fig. 1A). To further evaluate the differential expression of VDR , we compared their expression levels in the TCGA database using the platforms of GEPIA and UALCAN. The results showed that the expression of VDR in breast cancer tissues is significantly higher than in normal tissues (P < 0.01) (Fig. 1B, C).
As shown in Fig. 2, the prognostic value of VDR expression in breast cancer was analyzed using the Kaplan-Meier plotter, GEPIA, and UALCAN platforms. The results of the Kaplan-Meier plotter analysis indicated that overexpression of VDR was significantly associated with worse overall survival (OS) in breast cancer (P < 0.001) (Fig. 2A). Analysis of the relationship between VDR expression and OS in breast cancer patients by GEPIA and UALCAN platforms showed that overexpression of VDR was related to poor prognosis (P < 0.05) (Fig. 2B, C).
We plotted the Kaplan-Meier survival curves for molecular subtypes using the web-based curator. In the luminal A, luminal B, triple-negative breast cancer (TNBC), and human epidermal growth factor receptor 2 (HER2) subtypes of breast cancer, lower VDR expression of VDR was associated with worse prognosis (Fig. 3). This association was statistically significant. This analysis found that high expression of VDR in these molecular subtypes of breast cancer is associated with unfavorable prognostic factors and a high risk of breast cancer death. Hence, high VDR expression is a negative prognostic factor.
Identifying the co-expressed genes would improve the understanding of potential functions of VDR in breast cancer. Thus, the co-expressed genes of VDR were predicted by using TCGA database and GEO database (Fig. 4A, B); and, finally identified 960 DEGs (Fig. 4C). With the cutoff criteria of P < 0.01 and |logFC| > 1.5, we identified 423 DEGs from TCGA and GEO using 2 datasets (GSE27155, GSE35570). The 960 DEGs common between the GEO and TCGA databases were further used for pathway enrichment analysis.
A gene involved in a signaling pathway is normally co-expressed with other genes; thus, various genes collectively play a significant role in human cancer. Here, we identified genes whose expression correlates with that of VDR in breast cancers. We performed GO analysis of candidate DEGs using an online database. The DEGs were classified into the BP, CC, and MF groups (Fig. 5A). In the BP group, DEGs were mainly enriched in metabolic process and CC organization or biogenesis. In the CC group, DEGs were mainly enriched in the endomembrane system and extracellular space. In the MF group, DEGs were mainly enriched in protein binding and ion binding. According to the KEGG pathway enrichment analysis, DEGs were significantly enriched in pathways in cancer, malaria, extracellular matrix-receptor interactions, PI3K-AKT signaling pathway, and proteoglycans in cancer. DEGs were also enriched in complement and coagulation cascades and tyrosine metabolism (Fig. 5B).
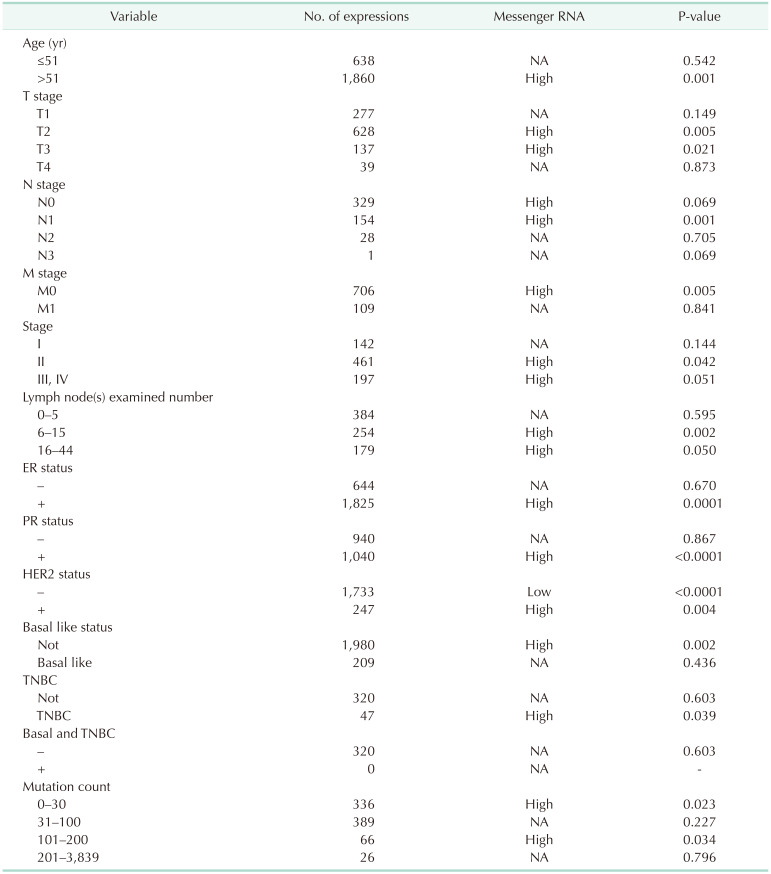
In the TCGA database, the expression of VDR was correlated with clinicopathological features of breast cancer patients (Fig. 6). The expression of VDR was related to estrogen receptor (ER) status, progesterone receptor (PR) status, HER2 status, basallike PAM 50 status, TNBC status, and basal-like PAM 50 & TNBC status (P < 0.05). We further investigated the associations between VDR expression and different clinicopathological characteristics in patients with breast cancer. As shown in Table 1, among breast cancer patients, VDR expression was higher in the >51-year-old group than in the ≤51-year-old group. In different TNM stages, VDR was significantly expressed in the T2 and T3 stages. VDR was the high expression in the N0 and N1 stages, and VDR was the high expression in the M0 stage. In different stages, VDR had significantly higher expression in stages II, III, and IV. The VDR was found to be highly expressed in both ER-positive and PR-positive statuses. Interestingly, VDR showed clinical significance in both HER2 negative and positive breast cancer patients. The expression of VDR was significantly upregulated in non-basal-like statues and triple-negative subtypes. In addition, high VDR expression also tended to correlate with mutation counts of 0–30 (Table 1).
Fig. 7A shows VDR expression according to histological type and Fig. 7B according to molecular subtype. The VDR mRNA level was lower in luminal A type tumors than in HER2, and in luminal B than in basal-like and HER2 and was higher in HER2 type than in basal-like; all these differences were statistically significant.
The PPI network of VDR was generated using the STRING database. The result showed that the following proteins showed interactions with VDR: receptor for retinoic acid (RXRA), E1A-binding protein P300 (EP300), nuclear receptor coactivator 1 (NCOA1), CREB-binding protein (CREBBP), space mission analysis and design (SMAD)-3, SMAD4, retinoid X receptor beta (RXRB), mediator of RNA polymerase II transcription subunit 1 (MED1), bromodomain adjacent to zinc finger domain 1B (BAZ1B), and retinoid X receptor gamma (RXRG) (Fig. 8A). To identify the hub genes, we used the CytoHubba Cytoscape plugin and extracted data from 4 calculation methods (edge percolated component, maximum cluster centrality, maximum neighborhood component, and degree). We selected the top 5 nodes ranked by these 4 methods. The top 5 genes were VDR, RXRA, NCAO1, EP300, and CREBBP (Fig. 8B). Thus, these predicted interacting partners of VDR may be involved in the regulation of VDR-mediated cancer progression and prognosis.
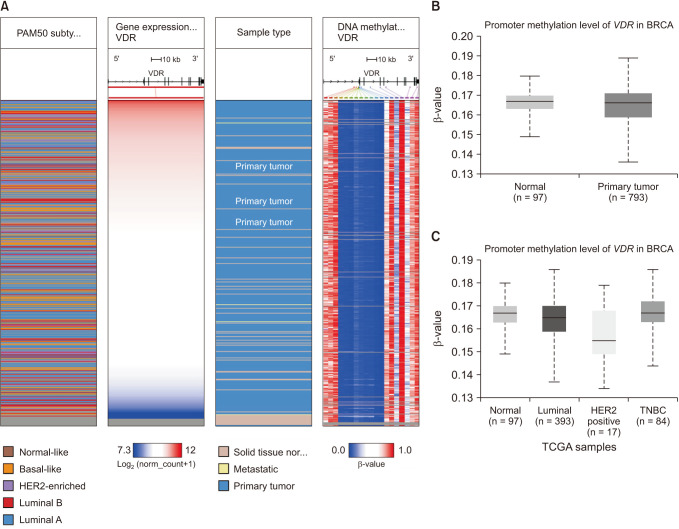
We also investigated the methylation status of the promoter of the VDR gene using UCSC Xena, an interactive web viewer for the visualization of DNA methylation based on TCGA data (Fig. 9A). We found that the VDR promoter is significantly hypermethylated in breast cancer tissue compared to the corresponding normal tissue (Fig. 9B). We also found that it is hypermethylated in TNBC, whereas the level of methylation is significantly lower in HER2+ (Fig. 9C).
Breast cancer is the first or second most common cancer among females worldwide. Many studies have been conducted to assess the relationship between the VDR gene and breast cancer development [21]. The purpose of this study was to examine the role of VDR in the prognosis of breast cancer. We conducted a bioinformatics analysis of various gene expression datasets with clearly defined distinguishing parameters between cancer and normal tissues. We found that VDR is expressed differently in cancer and normal tissues. According to our Oncomine, GEPIA, and UALCAN analysis, VDR is upregulated in lymphoma, glioblastoma, rectum adenocarcinoma, myeloma and breast, lung, and ovarian cancers, but downregulated in colon, esophageal, gastric, kidney, and prostate cancers. The expression of VDR in breast cancer was significantly higher than that in normal breast tissue.
In previous research, VDR expression was found to be associated with favorable prognostic characteristics, such as small size, low grade, ER positivity, PR positivity, low Ki67 expression, and luminal-like molecular subtypes [2223]. In these studies, ER and HER2 status was found to be positively correlated with VDR expression. Conversely, basal-like status and TNBC status were negatively correlated with VDR expression.
We further investigated the prognostic value of VDR in breast cancer using Kaplan-Meier plotter, and the pooled results revealed that lower VDR expression correlated with shorter OS, RFS, distant metastasis-free survival, and postprogression survival. Together, these findings indicate that increased expression of VDR might be a useful marker in the prognosis of breast cancer. A previous study [22] has associated high expression of VDR in invasive breast tumors with favorable prognostic factors and a low risk of breast cancer death. Hence, high VDR expression appears to be a positive prognostic factor [22]. In contrast, our results indicate that high VDR expression was associated with poor survival in breast cancer. Thus, because of the contradictory results from different analyses, its role in breast cancer remains unclear.
Tumorigenesis is a multi-step process that leads to tumor growth [24]. It is affected primarily by 4 major factors: somatically acquired genetic, epigenetic, transcriptomic, and proteomic alterations [24]. Genomic regions with possible inhibitory or carcinogenic effects undergo simultaneous loss-of-function and gain-of-function effect [2425]. Therefore, we used cBioPortal to identify breast cancer with copy number alterations and mutations in VDR genes. Missense and truncating mutations predominantly occurred within protein-coding sequences of these genes.
Through co-expression and correlation analysis, we observed that ASAP1 expression was positively correlated with VDR expression. ASAP1 is a multi-domain member of the ARFGAP protein family and has roles in metastasis in several systems including breast cancer cell lines, in which it has been implicated in invasion and metastatic potential [26]. Co-expression of these 2 genes may have implications for clinical outcomes in patients with breast cancer.
We then used the online tool to conduct GO and KEGG pathway analyses to evaluate the common role of these associated genes in breast cancer. VDR-correlated genes demonstrated more varied characteristics than VDR-correlated genes (12 vs. 31 pathways). This analysis indicated that VDR performs various functions with regard to pathway regulation; however, they may have some similar functions in breast cancer signaling pathways and in breast cancer types. The VDR promoter is hypermethylated in breast cancer, its demethylation in breast cancer cell lines results in reexpression of VDR transcripts [23]. Similar epigenetic suppression of VDR mRNA expression has been demonstrated in placental and choriocarcinoma cell lines, which also show VDR mRNA reexpression after azacitidine and its deoxy derivative, decitabine (azacitidine) treatment [2327]. Comparison of the VDR expression heat map and DNA methylation status revealed that its expression might be negatively related to some CpG sites (blank frame).
In conclusion, the present study investigated the functions of VDR as predictive biomarkers in breast cancer using a comprehensive multiomic approach. Although we explored the relationship between VDR and breast cancer patients, there are still certain restrictions necessitating more study. Nevertheless, our research provides suggestions concerning the predictive function of VDR in breast cancer.
Notes
References
1. DeSantis C, Siegel R, Bandi P, Jemal A. Breast cancer statistics, 2011. CA Cancer J Clin. 2011; 61:409–418. PMID: 21969133.

2. Petracci E, Decarli A, Schairer C, Pfeiffer RM, Pee D, Masala G, et al. Risk factor modification and projections of absolute breast cancer risk. J Natl Cancer Inst. 2011; 103:1037–1048. PMID: 21705679.

3. Giovannucci E. The epidemiology of vitamin D and cancer incidence and mortality: a review (United States). Cancer Causes Control. 2005; 16:83–95. PMID: 15868450.

4. Keum N, Giovannucci E. Vitamin D supplements and cancer incidence and mortality: a meta-analysis. Br J Cancer. 2014; 111:976–980. PMID: 24918818.

5. Margolis RN, Christakos S. The nuclear receptor superfamily of steroid hormones and vitamin D gene regulation: an update. Ann N Y Acad Sci. 2010; 1192:208–214. PMID: 20392238.

6. Deeb KK, Trump DL, Johnson CS. Vitamin D signalling pathways in cancer: potential for anticancer therapeutics. Nat Rev Cancer. 2007; 7:684–700. PMID: 17721433.

7. Feldman D, Krishnan AV, Swami S, Giovannucci E, Feldman BJ. The role of vitamin D in reducing cancer risk and progression. Nat Rev Cancer. 2014; 14:342–357. PMID: 24705652.

8. Welsh J. Vitamin D and breast cancer: insights from animal models. Am J Clin Nutr. 2004; 80(6 Suppl):1721S–1724S. PMID: 15585794.

9. Anzano MA, Smith JM, Uskoković MR, Peer CW, Mullen LT, Letterio JJ, et al. 1 alpha,25-Dihydroxy-16-ene-23-yne-26,27-hexafluorocholecalciferol (Ro24-5531), a new deltanoid (vitamin D analogue) for prevention of breast cancer in the rat. Cancer Res. 1994; 54:1653–1656. PMID: 8137276.
10. Crew KD, Gammon MD, Steck SE, Hershman DL, Cremers S, Dworakowski E, et al. Association between plasma 25-hydroxyvitamin D and breast cancer risk. Cancer Prev Res (Phila). 2009; 2:598–604. PMID: 19470790.

11. Murray A, Madden SF, Synnott NC, Klinger R, O’Connor D, O’Donovan N, et al. Vitamin D receptor as a target for breast cancer therapy. Endocr Relat Cancer. 2017; 24:181–195. PMID: 28213567.

12. Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004; 6:1–6. PMID: 15068665.

13. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017; 45(W1):W98–W102. PMID: 28407145.

14. Chandrashekar DS, Bashel B, Balasubramanya SA, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BV, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017; 19:649–658. PMID: 28732212.

15. Gross MI, Demo SD, Dennison JB, Chen L, Chernov-Rogan T, Goyal B, et al. Antitumor activity of the glutaminase inhibitor CB-839 in triple-negative breast cancer. Mol Cancer Ther. 2014; 13:890–901. PMID: 24523301.

16. Jézéquel P, Campone M, Gouraud W, Guérin-Charbonnel C, Leux C, Ricolleau G, et al. bc-GenExMiner: an easy-to-use online platform for gene prognostic analyses in breast cancer. Breast Cancer Res Treat. 2012; 131:765–775. PMID: 21452023.

17. Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017; 45(D1):D353–D361. PMID: 27899662.

18. Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017; 45(D1):D362–D368. PMID: 27924014.

19. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003; 13:2498–2504. PMID: 14597658.

20. Goldman M, Craft B, Zhu J, Haussler D. UCSC Xena for cancer genomics visualization and interpretation. Cancer Res. 2019; 79(13 Suppl):Abstract nr 911.
21. Shaikh F, Baig S, Jamal Q. Do VDR gene polymorphisms contribute to breast cancer? Asian Pac J Cancer Prev. 2016; 17:479–483. PMID: 26925631.

22. Huss L, Butt ST, Borgquist S, Elebro K, Sandsveden M, Rosendahl A, et al. Vitamin D receptor expression in invasive breast tumors and breast cancer survival. Breast Cancer Res. 2019; 21:84. PMID: 31358030.

23. Al-Azhri J, Zhang Y, Bshara W, Zirpoli G, McCann SE, Khoury T, et al. Tumor expression of vitamin D receptor and breast cancer histopathological characteristics and prognosis. Clin Cancer Res. 2017; 23:97–103. PMID: 27407090.

24. Chakravarthi BV, Nepal S, Varambally S. Genomic and epigenomic alterations in cancer. Am J Pathol. 2016; 186:1724–1735. PMID: 27338107.

25. Herceg Z, Hainaut P. Genet ic and epigenetic alterations as biomarkers for cancer detection, diagnosis and prognosis. Mol Oncol. 2007; 1:26–41. PMID: 19383285.

26. Onodera Y, Hashimoto S, Hashimoto A, Morishige M, Mazaki Y, Yamada A, et al. Expression of AMAP1, an ArfGAP, provides novel targets to inhibit breast cancer invasive activities. EMBO J. 2005; 24:963–973. PMID: 15719014.

27. Pospechova K, Rozehnal V, Stejskalova L, Vrzal R, Pospisilova N, Jamborova G, et al. Expression and activity of vitamin D receptor in the human placenta and in choriocarcinoma BeWo and JEG-3 cell lines. Mol Cell Endocrinol. 2009; 299:178–187. PMID: 19133314.

Fig. 1
Expression of VDR messenger RNA (mRNA) in cancer vs. normal (Oncomine and TCGA database). (A) This graphic generated by Oncomine (https://www.oncomine.org) indicates the number of datasets with statistically significant (P < 0.01) mRNA overexpression (red) or down expression (blue) of VDR (different types of cancer vs. corresponding normal tissue). The threshold was designed with the following parameters: P-value of 1e-4, fold change of 2, and gene ranking of 10%. (B) The expression of VDR in breast cancer tissue and normal breast tissue generated by GEPIA 2 web (http://gepia2.cancer-pku.cn/#index) (TCGA data), P < 0.01. (C) The expression of VDR in breast cancer based on sample types by UALCAN (http://ualcan.path.uab.edu/index.html), P < 0.001. TCGA, The Cancer Genome Atlas; BRCA, breast cancer.
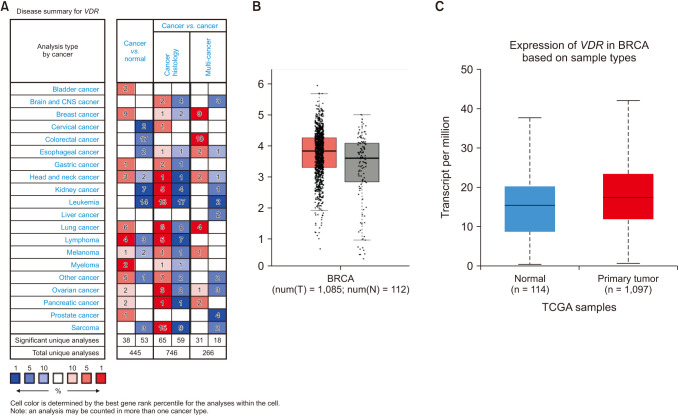
Fig. 2
Survival of VDR expression in breast cancer. (A) The survival curve comparing the breast cancer patient with the high (red) and low (black) expression of VDR was plotted using the Kaplan-Meier plotter (https://kmplot.com/analysis/) (P < 0.001). (B) The survival curve comparing the breast cancer patient with the high (red) and low (blue) expression of VDR was plotted using GEPIA (http://gepia2.cancer-pku.cn/#survival) (P < 0.05). TPM, transcripts per kilobase per million mapped; HR, hazards ratio; P, P-value; n, number. (C) The survival curve comparing the breast cancer patient with the high (red) and low (blue) expression of VDR was plotted using UALCAN (http://ualcan.path.uab.edu/index.html) (P < 0.05).
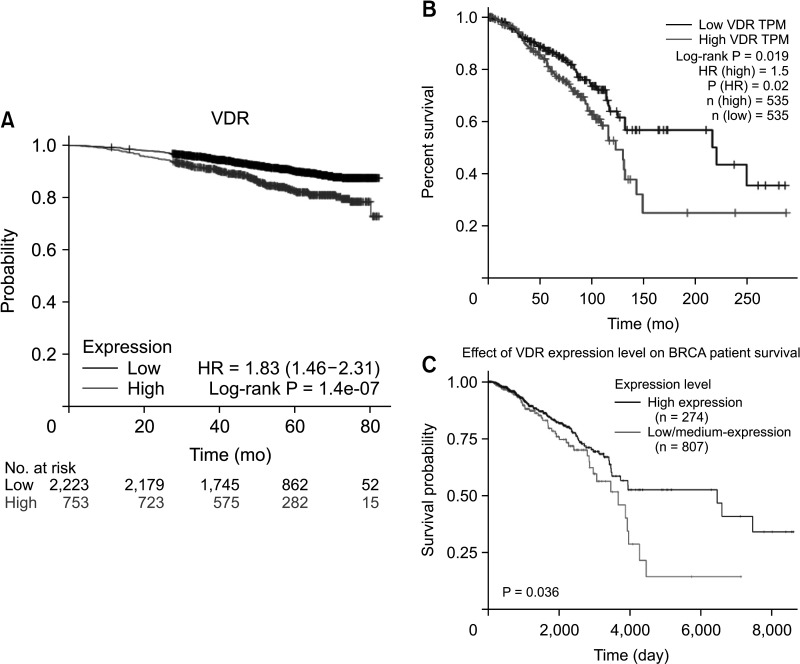
Fig. 3
Molecular subtypes prognostic analysis generated by bc-GenExMiner (breast cancer Gene-Expression Miner) ver. 4.2 (http://bcgenex.centregauducheau.fr/BC-GEM/GEM-requete.php?mode=8). (A) Luminal A, (B) luminal B, (C) triple-negative breast cancer (TNBC), and (D) human epidermal growth factor receptor 2+ (HER2+). OS, overall survival; HR, hazard ratio; CI, confidence interval; DMFS, distant metastasis-free survival.
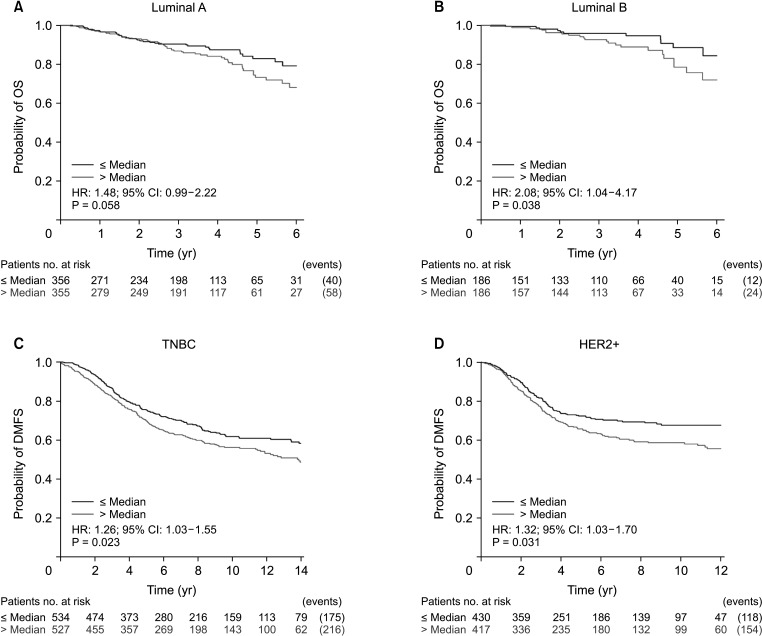
Fig. 4
Co-expression analysis of VDR gene expression. (A) VDR co-expression of genes analyzed using TCGA (The Cancer Genome Atlas) data. (B) VDR co-expression of genes analyzed using GEO (Gene Expression Omnibus) (GSE7849). (C) The intersection genes of VDR co-expression.
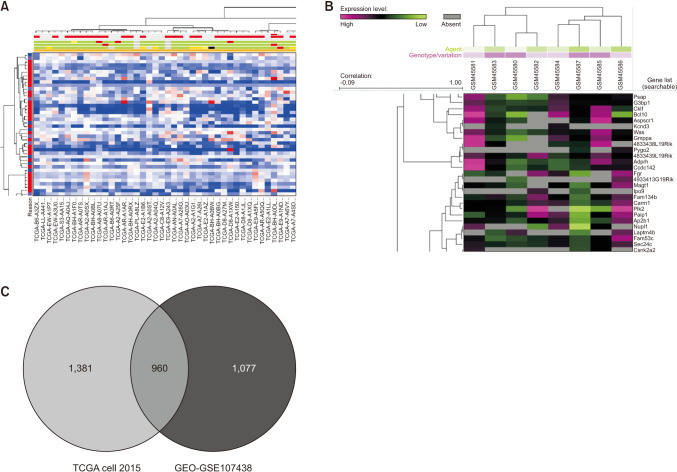
Fig. 5
(A) Gene ontology (GO) analysis of VDR. (B) Kyoto Encyclopedia of Genes and Genome (KEGG) pathway enrichment analysis of VDR. The functional enrichment analysis of VDR using the web tool WebGestalt (WEB-based Gene SeT AnaLysis Toolkit) (http://www.webgestalt.org/). ECM, extracellular matrix.
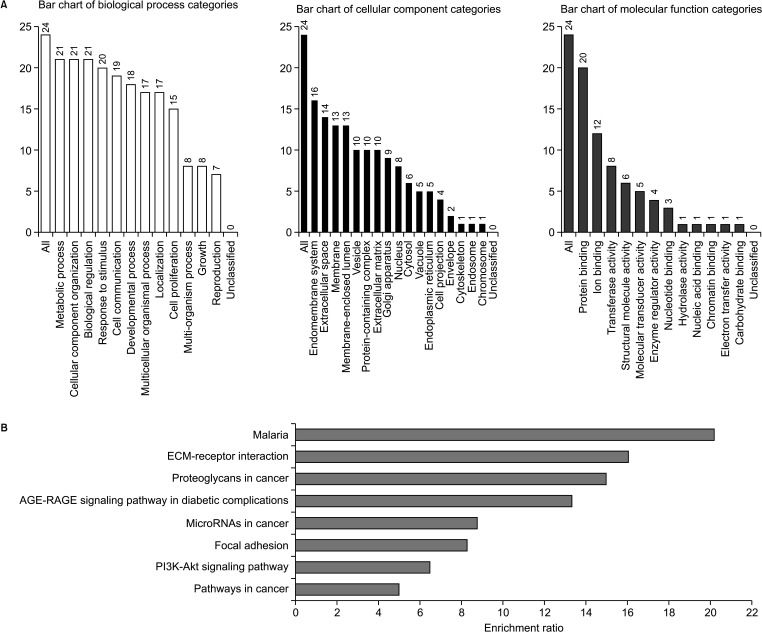
Fig. 6
The relationship between expression of VDR and various clinicopathological parameters of breast cancer using bc-GenExMiner (breast cancer Gene-Expression Miner) ver. 4.2 (http://bcgenex.centregauducheau.fr/BC-GEM/GEM-requete.php). ER, estrogen receptor; IHC, immunohistochemistry; mRNA, messenger RNA; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2; TNBC, triple-negative breast cancer.
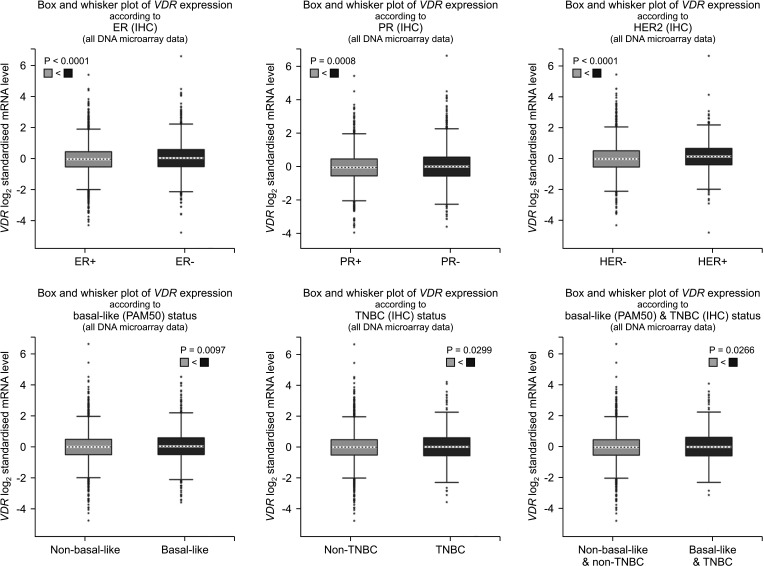
Fig. 7
The expression of VDR in breast cancer by histological and molecular subtypes using bc-GenExMiner (breast cancer Gene-Expression Miner) ver. 4.2 (http://bcgenex.centregauducheau.fr/BC-GEM/GEM-requete.php). (A) The VDR expression of histological type. (B) The VDR expression of molecular subtypes. mRNA, messenger RNA; HER2, human epidermal growth factor receptor 2.
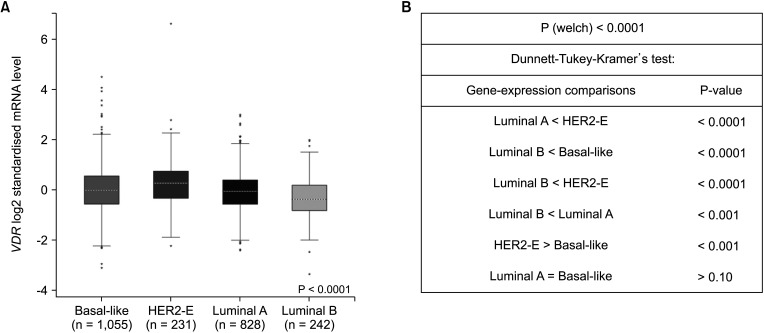
Fig. 8
Identification of known and predicted structural proteins essential for VDR function. (A) The protein-protein interaction network of VDR was generated using STRING database (https://string-db.org). (B) Top 5 hub genes were identified using the CytoHubba Cytoscape plugin with extracted data from degree of calculation methods.
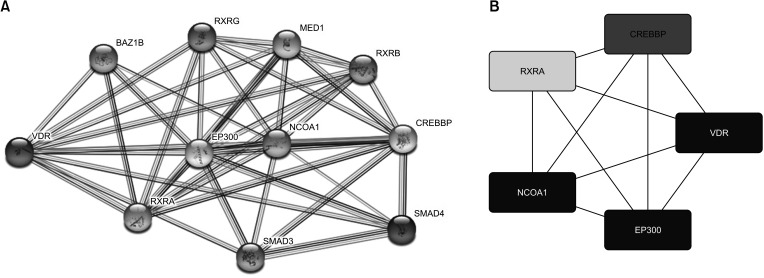
Fig. 9
Correlation between VDR messenger RNA expression and promoter methylation in breast cancer (TCGA data). (A) Heat map of VDR expression and DNA methylation status retrieved from UCSC Xena web (https://xenabrowser.net/heatmap/). (B) The promoter methylation level of the VDR gene in breast cancer tissue compared to their normal tissue. (C) The promoter methylation level of the VDR gene in breast cancer stages. The box plot was retrieved from UALCAN web (http://ualcan.path.uab.edu). TCGA, The Cancer Genome Atlas; BRCA, breast cancer; HER2, human epidermal growth factor receptor 2; TNBC, triple-negative breast cancer.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download