INTRODUCTION
METHODS
Reagents
Establishment of a rat model of DN
H&E staining
Measurement of related index parameters
Cell culture and treatment
PI staining
ELISA
Western blotting
RT-qPCR
Table 1
Statistical analysis
RESULTS
Pyroptosis in DN rat kidney tissues and HK-2 cells
 | Fig. 1Changes in the kidney function and morphology in diabetic rats.A rat model wherein streptozotocin (STZ) was used to induce diabetic nephropathy (DN). (A) H&E staining to observe the morphological characteristics of the kidney (scale bar 50 μm, ×400; scale bar 200 μm, ×100) magnifications, (B) Albumin/creatinine ratio. (C) Plasma cystatin-C levels. (D) Glomerular volume. (E) Mesangial area. (F) Tuft area. Data are presented as mean ± SEM from 6 rats. Compared to control, **p < 0.01.
|
 | Fig. 2In vivo and in vitro expression of pyroptosis-related proteins.(A) Bands corresponding to the expression of pyroptosis-related proteins (caspase-1 p45, cleaved caspase-1, interleukin [IL]-1β, IL-18, gasdermin D [GSDMD], and GSDMD-N) in the kidneys of diabetic nephropathy (DN) rats. (B) Quantification of the bands observed in (A). (C) Concentration of IL-1β in rat plasma. (D) Concentration of IL-18 in rat plasma. (E) Bands corresponding to the expression of pyroptosis-related proteins (caspase-1 p45, cleaved caspase-1, IL-1β, IL-18, GSDMD, and GSDMD-N) in high glucose (HG)-treated HK-2 cells. (F) Quantification of the bands observed in (E). (G) Concentration of IL-1β in HK-2 cell supernatant. (H) Concentration of IL-18 in HK-2 cell supernatant. Data are presented as mean ± SEM from 6 rats and 3 replicated cell experiments. Compared to control, *p < 0.05, **p < 0.01.
|
In vivo and in vitro investigation of the role of DDR1 in pyroptosis
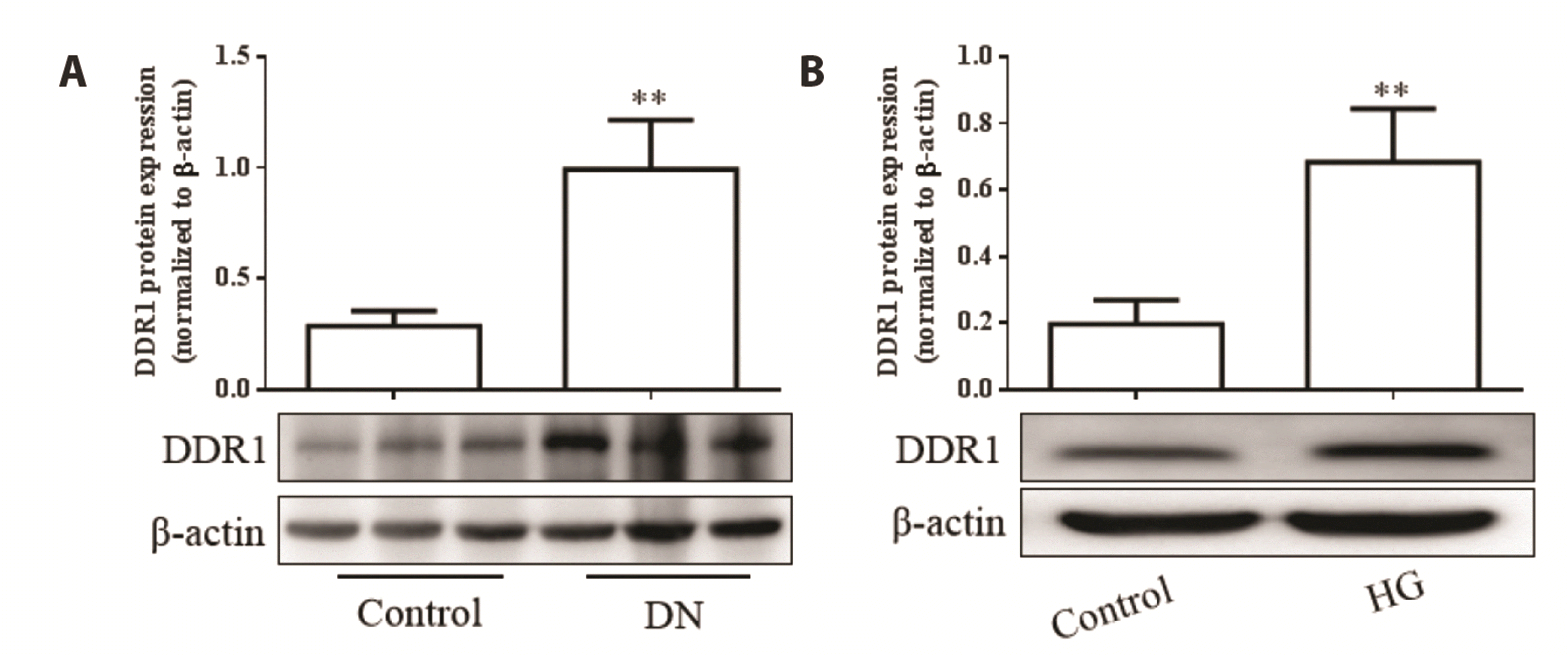 | Fig. 3In vivo and in vitro expression of discoid domain receptor 1 (DDR1).(A) DDR1 expression in the kidneys of diabetic nephropathy (DN) rats. (B) DDR1 expression in high glucose (HG)-treated HK-2 cells. Data are presented as mean protein in the kidneys of DN rats and HG-treated HK-2 cells. Data are presented as mean ± SEM from 6 rats and 3 replicated cell experiments. Compared to control, **p < 0.01.
|
Role of DDR1 in pyroptosis of HG-treated HK-2 cells
 | Fig. 4Effect of discoid domain receptor 1 (DDR1) knockdown on HK-2 cell pyroptosis.(A) Bands corresponding to the expression of DDR1 in the background of transfection with siDDR1. (B) Quantification of the band observed in (B). (C) Propidium iodide (PI) staining was used to analyze HK-2 cell death (scale bar: 200 μm). (D) Percentage HK-2 cell pyroptosis. (E) Bands corresponding to the expression of pyroptosis-related proteins (caspase-1 p45, cleaved caspase-1, interleukin [IL]-1β, IL-18, gasdermin D [GSDMD], and GSDMD-N). (F) Quantification of the bands observed in (E). (G) The concentration of IL-1β in HK-2 cell supernatant. (H) The concentration of IL-18 in HK-2 cell supernatant. Data are presented as mean ± SEM from 3 replicated cell experiments. Compared to high glucose (HG), *p < 0.05, **p < 0.01.
|
Role of DDR1 in NF-κB/NLRP3-mediated pyroptosis
 | Fig. 5Discoid domain receptor 1 (DDR1) knockdown resulted in the downregulation of the nuclear transcription factor-κB (NF-κB)/NLR family pyrin domain-containing 3 (NLRP3) pathway intermediaries at the protein level.(A) Bands corresponding to the expression of phosphorylated NF-κB (p-NF-κB) and NF-κB in the kidneys of DN rats. (B) Bands corresponding to the expression of NLRP3 in the kidneys of diabetic nephropathy (DN) rats. (C) Bands corresponding to the expression of p-NF-κB and NF-κB in HK-2 cells. (D) Bands corresponding to the expression of NLRP3 in HK-2 cells. Data are presented as mean ± SEM from 6 rats and 3 replicated cell experiments. Compared to control, **p < 0.01. Compared to high glucose (HG), #p < 0.05, ##p < 0.01.
|
Role of NLRP3 in DDR1-induced pyroptosis
 | Fig. 6NLR family pyrin domain-containing 3 (NLRP3) reversed the discoid domain receptor 1 (DDR1) knockdown‒mediated downregulation of the nuclear transcription factor-κB (NF-κB)/NLRP3 pathway intermediaries at the transcript level (assessed using RT-qPCR).Transcript-level expression of (A) NLRP3, (B) NF-κB, (C) caspase-1, (D) gasdermin D (GSDMD), (E) interleukin (IL)-1β, (F) IL-18. Data are presented as mean ± SEM from 3 replicated cell experiments. Compared to control **p < 0.01. Compared to high glucose (HG), ##p < 0.01. Compared to siDDR1, &&p < 0.01.
|




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download