Pseudomonas aeruginosa is the etiologic agent of various nosocomial infections, such as sepsis, pneumonia, and urinary tract infections. The majority of
P. aeruginosa isolates are resistant to carbapenems [
1]. Carbapenem resistance is frequently associated with loss of the outer membrane porin OrpD or expression of the efflux system, combined with extended-spectrum β-lactase production or AmpC hyperproduction [
2]. Another important antimicrobial resistance mechanism is the acquisition of carbapenemase genes, such as those encoding Amber class A (GES type) and class B metallo-β-lactamases, including imipenemase (IMP), Verona integron metallo beta-lactamase (VIM), and New Delhi metallo- β-lactamase (NDM) [
3]. IMP-6–producing
P. aeruginosa sequence type (ST) 235 is the dominant clone of carbapenemase-producing
P. aeruginosa (CPPAE) in Korea [
4-
7]. A 2020 study by the Antimicrobial Resistance Surveillance System in Korea (Kor-GLASS) [
8] demonstrated an increase in the carbapenem resistance rate and a shift in the molecular epidemiology of
P. aeruginosa blood isolates in 2020. In this report, we summarize the dominant clones and their characteristics.
A total of 212 non-duplicated
P. aeruginosa blood isolates were isolated according to the Kor-GLASS manual [
8]; the isolates were collected from nine general hospitals (three general hospitals in Seoul or Gyeonggi-do, two general hospitals in Gyeongsang-do, one in Gangwon-do, one in Jeolla-do, one in Chungcheong-do, and one in Jeju-do) and two nursing homes in Seoul. Of these, 24 isolates were identified as CPPAE. Due to the purely observational nature and very low risk to individual privacy of the participants, this study was approved by local institutional review boards (approval number: NHIMC-2022-06-012) and exempted from the requirement of informed consent.
Pure colonies of
P. aeruginosa were collected in 10% skim milk and stored at −70°C before all collected isolates were transferred to the Korea Centers for Disease Control and Prevention (KCDC) analysis center using approved methods [
8]. Bacterial species were verified using matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (Bruker Biotyper, Bruker Daltonics GmbH, Bremen, Germany). Antimicrobial susceptibility was mainly determined by the disk diffusion test, except for the colistin minimum inhibitory concentration (MIC), which was determined by the broth microdilution method. The MICs of carbapenems (imipenem, meropenem, and doripenem) were also determined. The interpretation followed the CLSI guidelines [
9].
P. aeruginosa isolates showing no susceptibility to imipenem or meropenem were PCR-sequenced to detect
blaKPC,
blaNDM,
blaOXA-48,
blaVIM,
blaIMP, and
blaGES. DNA of freshly subcultured isolates was extracted using GenElute Bacterial Genomic DNA Kit (Sigma-Aldrich, St. Louis, MO, USA). Genomic DNA concentration was measured using Qubit analyses (Thermo Fisher Scientific, Waltham, MA, USA), and 8 μg of input DNA was used. The entire genomes of the CPPAE isolates were sequenced using a NextSeq 550 instrument (Illumina, San Diego, CA, USA), and sequences were assembled using Spades (version 3.11.1) and annotated using Prokka (version 1.13.7). Resistance genes were obtained from the Center for Genomic Epidemiology website with ResFinder 4.1 [
10]. Multilocus sequence typing (MLST) was determined with the following seven housekeeping genes: acetyl coenzyme A synthetase (
acsA), Shikimate dehydrogenase (
aroE), GMP synthase (
guaA), DNA mismatch repair protein (
mutL), NADH dehydrogenase I chain C, D (
nuoD), phosphoenolpyruvate synthase (
ppsA), and anthranilate synthetase component I (
trpE) from the Center for Genomic Epidemiology [
10].
In this study, resistance to carbapenem in
P. aeruginosa blood isolates were concerning with the resistance rate of approximately 35% (
Fig. 1). Especially, all CPPAE isolates were resistant to gentamicin, tobramycin, and ciprofloxacin. Most common metallo-β-lactamase, detected in carbapenem-resistant
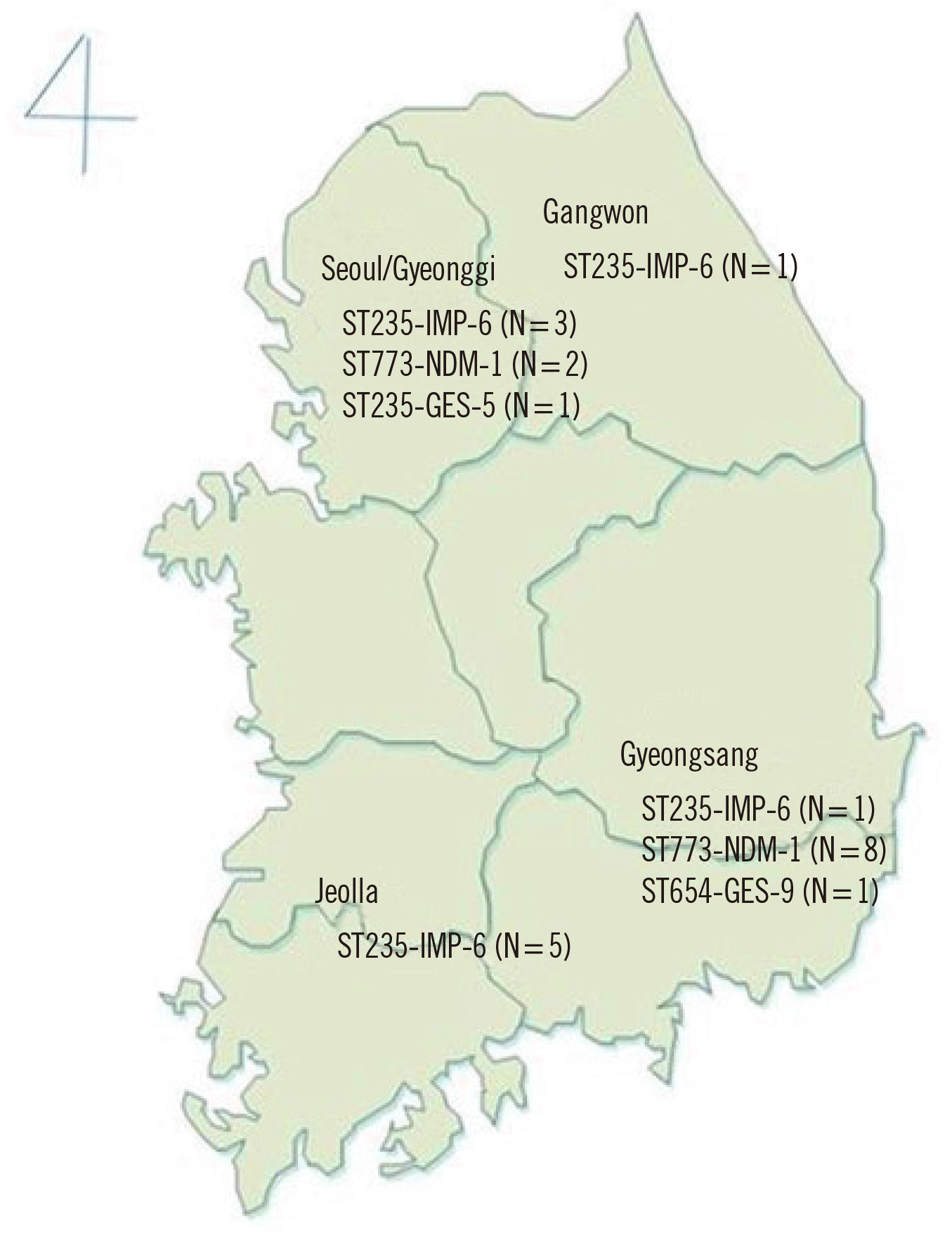
P. aeruginosa isolates was IMP-6 (N=11), following NDM-1 (N=10). Interestingly, all NDM-1–producing
P. aeruginosa strains were ST 773, where eight of them were isolated from two general hospitals in Gyeongsang-do (
Fig. 2). The CPPAE isolates had various antimicrobial resistance genes, which are summarized in
Supplemental Data Table 1.
Fig. 1
Antimicrobial resistance rates of P. aeruginosa blood isolates (N=212).
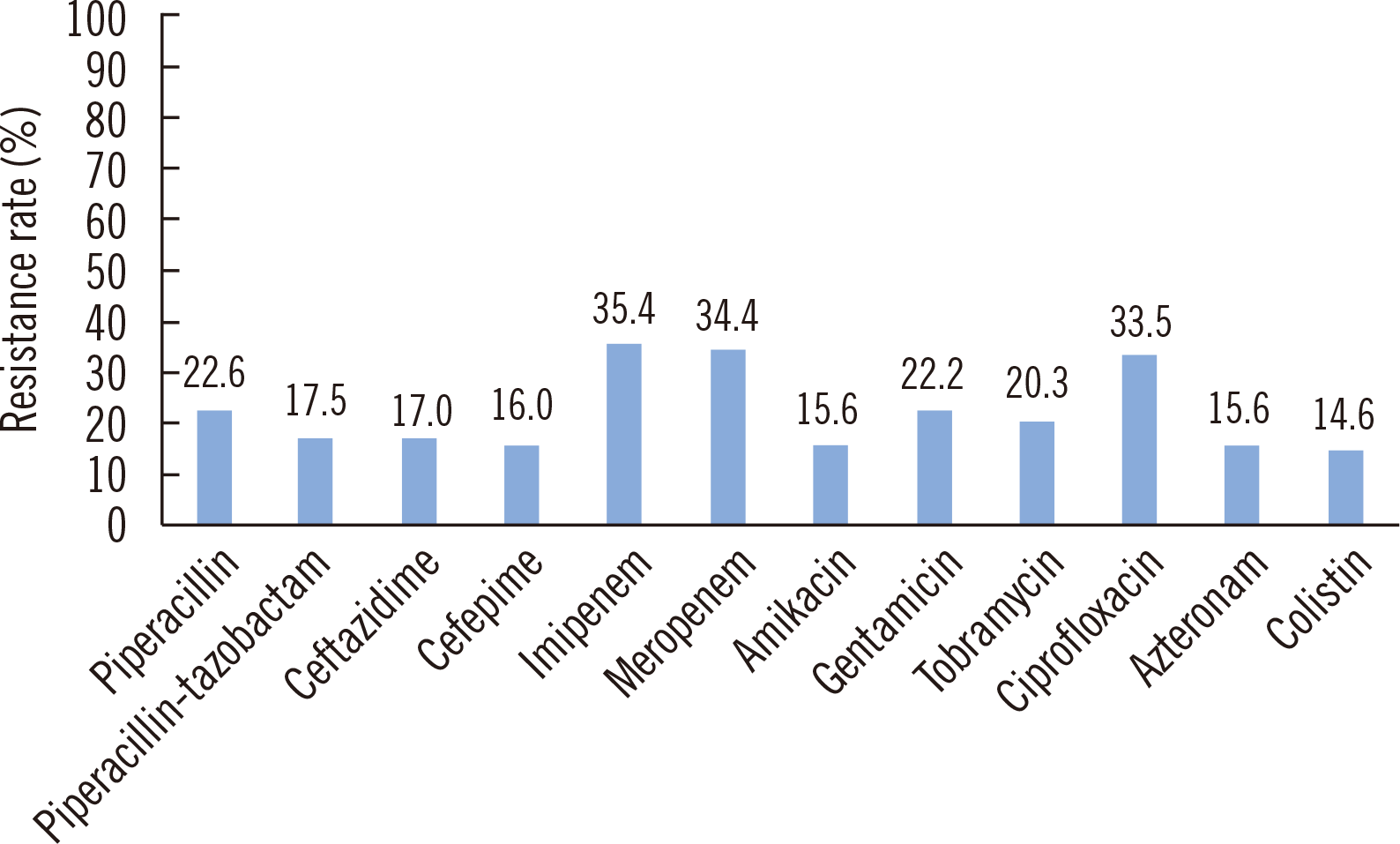

Fig. 2
Molecular epidemiology of carbapenemase-producing Pseudomonas aeruginosa isolates (sequence type: carbapenemase genotype). Carbapenemase-producing P. aeruginosa isolates were collected from two nursing homes in Seoul, three general hospitals in Seoul or Gyeonggi-do, one general hospital in Gangwon-do, one general hospital in Jeolla-do, and two general hospitals in Gyeongsang-do. Two isolates with undetermined sequence types were not shown in this map.

The increase in carbapenem-resistant
P. aeruginosa has become problematic considering the limitations of treatment options. Antimicrobial resistance due to carbapenemase genes is more challenging than that of other mechanisms such as membrane impermeability, with the possibility of horizontal gene transfer. Nationwide monitoring of the molecular epidemiology of CPPAE is helpful in establishing an adequate strategic policy for the control of antimicrobial resistance. IMP-6–producing
P. aeruginosa ST 235 is the dominant clone in Korea [
4-
7]. IMP-1 is a globally prevalent subtype among 79 IMP variants, whereas IMP-6 is the dominant subtype in Korea [
3]. IMP-6 has better hydrolyzing activity against meropenem. The frequent use of meropenem in clinical settings may have contributed to the spread of this type in Korea [
3]. GES-type CPPAE clinical isolates from long-term care facilities and general hospitals are limited in Korea [
11].
A recent study reported the clonal spread of NDM-1–producing
P. aeruginosa ST 773 isolates possessing
rmtB4, mostly from urine isolates, at a university hospital in Korea [
12]. In the present nationwide surveillance of blood isolates, the emergence of NDM-1–producing
P. aeruginosa ST 773 was found mostly in Gyeongsang Province, which may indicate a shift in the molecular epidemiology of CPPAE. NDM-producing
P. aeruginosa has been identified mostly in Asia, Europe, and Africa, demonstrating intercontinental dissemination. NDM-1 is the most prevalent subtype, but it is rarely reported in Korea [
3]. Recently, NDM-1–producing
P. aeruginosa isolates were reported at a university hospital in Seoul, Korea [
12].
The combination of carbapenem and amikacin has become an important treatment option to combat multidrug-resistant or extensively drug-resistant
P. aeruginosa [
13]. In a previous report, NDM-1–producing
P. aeruginosa isolates had the highest sensitivity to tigecycline and amikacin [
14]. In the present study, high rates of resistance to amikacin were also observed for CPPAE. All the NDM-1–carrying
P. aeruginosa strains were resistant to amikacin and possessed
rmtB. Junaid [
14] also reported that NDM-producing
P. aeruginosa isolates co-harboring
rmtC showed a very high amikacin MIC (more than 2,048 μg/mL). Continuous surveillance is necessary to prevent the infection and transmission of carbapenem- and amikacin-resistant
P. aeruginosa in Korea.

