Abstract
Background
Most pain management techniques for challenging procedures are still performed under the guidance of the C-arm fluoroscope although it is sometimes difficult for even experienced clinicians to understand the modified three-dimensional anatomy as a two-dimensional X-ray image. To overcome these difficulties, the development of a virtual simulator may be helpful. Therefore, in this study, the authors developed a virtual simulator and presented its clinical application cases.
Methods
We developed a computer program to simulate the actual environment of the procedure. Computed tomography (CT) Digital Imaging and Communications in Medicine (DICOM) data were used for the simulations. Virtual needle placement was simulated at the most appropriate position for a successful block. Using a virtual C-arm, the authors searched for the position of the C-arm at which the needle was visualized as a point. The positional relationships between the anatomy of the patient and the needle were identified.
Results
For the simulations, the CT DICOM data of patients who visited the outpatient clinic was used. When the patients revisited the clinic, images similar to the simulated images were obtained by manipulating the C-arm. Transforaminal epidural injection, which was difficult to perform due to severe spinal deformity, and the challenging procedures of the superior hypogastric plexus block and Gasserian ganglion block, were successfully performed with the help of the simulation.
The development of image-processing technology in modern society has led to advancements in all aspects of diagnosis and treatment in medicine over the past few decades. Virtual simulation, in particular, has rapidly progressed in various fields of modern medicine by improving navigation and enabling visualization of the complex anatomy of a patient prior to surgery [1–3].
Despite the increasing variability and difficulty in performing procedures for pain intervention, many procedures are still performed under the guidance of the C-arm [4], which is a movable device that can generate X-rays in a desired direction and location. However, the images produced by the C-arm are two-dimensional (2-D) and, from a certain position, even experienced clinicians sometimes face difficulties performing procedures under C-arm guidance. When the anatomical structure of the patient is severely deformed or when the procedure itself is challenging, it becomes more difficult to identify the anatomical structures and position the needle at the desired target.
Several attempts have been made to perform the procedure more accurately and easily with the aid of virtual simulation [1–3]. Visualizing complex structures through virtual simulation may aid the clinicians in successfully performing procedures that are expected to be challenging. Although the creation of a virtual X-ray based on patient data has been introduced earlier [5], there are only a few studies in which this technique could be applied and utilized in clinical cases.
Therefore, a virtual simulation program was developed for use prior to challenging procedures. By generating a virtual X-ray, the authors aimed to overcome the difficult anatomy of the patient and difficulty in needle positioning. Herein is presented the development of a virtual simulation program using the Digital Imaging and Communications in Medicine (DICOM) image data of the patient as well as its clinical application.
Development of a program for the virtual simulation in this study was performed using the image data from the anonymized data server of Korea University Anam Hospital. The image data were obtained using the DICOM format. Each simulation was customized for each patient. Consent was obtained from all patients about using their computed tomography (CT) images to build their own simulation and to report and publish their simulation cases.
To obtain a 3-D polygonized model with a better visualization, segmentation was performed using the 3D Slicer version 4.10.2 (http://www.slicer.org).
For the vertebrae, 3-D data were obtained separately from each of the five lumbar vertebrae, the sacrum, and the pelvic bones. In addition, 3-D polygonal shapes of the intervertebral discs between each vertebra were obtained. In the case of the skull, 3-D data as a single bone was obtained, on the theory that the area to be treated will not be greatly affected by the movement of other bones. The whole-body contour was also obtained by leaving all the remaining values except for air. Although each polygonized data point was segmented separately, their spatial positional references were maintained to obtain a combined 3-D structure that could be reassembled.
The structure obtained had approximately 60,000 to 120,000 polygons per bone. However, when the number of polygons increases, all vertices, edges, and faces require computing resources in the program. Equipment that implements virtual reality needs to perform these calculations about 60 times per second, so in general, if the total number of polygons exceeds 1 million, performance may be affected. Therefore, there was a risk of performance degradation when used directly in a program. Therefore, using MeshMixer version 3.5 (Autodesk Inc., San Rafael, CA), a model was used that reduced the number of polygons by 5%.
For image acquisition, it was necessary to load the DICOM file format in the form of binary data that could be manipulated directly in the virtual space. In the case of CT, the DICOM file contains information such as the spatial location, size, and direction of each datum; therefore, it is highly effective if it can be used directly rather than transforming it into another form. It was necessary to place both the data and virtual needle directly in the virtual space to generate a virtual X-ray quickly and accurately. For this, existing libraries were sought out; however, none met the requirements. Therefore, a DICOM file handler compatible with the UNITY engine had to be created. A program was created that could read and manipulate DICOM files in the UNITY version 4.2 (Unity Technologies, San Francisco, CA) environment. Since the Grassroots DICOM version 3.0 (http://gdcm.sourceforge.net) library was not compatible with UNITY, CMake version 3.119.1 (Kitware, Inc., Clifton Park, NY), Simplified Wrapper and Interface Generator version 4.0.2 (http://www.swig.org/) were used to set an environment that could read and write DICOM data in a virtual environment for the UNITY and Visual Studio 2019 version 16.10.3 (Microsoft, Redmond, WA). In the development environment created, DICOM data were read as binary data and were classified with tags such as Image Type (Tag# 0008,0008), Image Orientation (Tag# 0020, 0037), Instance Number (Tag# 0020, 0013), Image Position (Tag# 0020, 0032), Pixel Spacing (Tag# 0028, 0030), Windows Center (Tag# 0028,1050), Windows Width (Tag# 0028,1051), Rescale Intercept (Tag# 0028, 1052), Rescale Slope (Tag# 0028, 1053), Pixel Data (Tag# 7FE0,0010), etc., and stored into a separate series of arrays.
The Hounsfield unit (HU) value of each image was obtained by adjusting the pixel data value of the extracted data with the rescale intercept and rescale slope values. The CT image was obtained by assembling the pixel colors and adjusting the obtained HU values using the Windows center and Windows width values. The images were positioned in the virtual space using the values of image orientation, image position, and pixel spacing (Fig. 1A). When only HU values between 200 and 400 (adjusted according to bone density) were visualized, a 3-D bone-like image was obtained (Fig. 1B).
The spatial coordinates of the four corners of the obtained images were stored in arrays. An X-ray generator and a detector were set in a virtual space to reproduce the actual procedure using the C-arm. The spatial coordinates of the four corners of the detector were divided by the desired resolution (x × y) to set targets for a virtual X-ray that started from the generator, as shown in Fig. 2A (resolution 5 × 5) and Fig. 2B (resolution 10 × 10). The generator and detector were programmed such that the position and direction could be adjusted similar to the actual use of the C-arm. Using a keyboard, horizontal direction keys were set to adjust the caudad to cephalad tilting, and vertical direction keys were set to adjust the left-to-right rotation. The point of the CT slice images where the collision occurred was calculated using the ratio of the dot product of each X-ray with the reference point of the obtained images. After the corresponding HU values were obtained, the sum of these values was calculated. By dividing this value by the total number of CT slices (the number of the slices ranged from 200 to 600 depending on the patient), the degree of absorption of X-rays was calculated, and virtual X-rays were obtained by arranging them sequentially by converting the values into pixel values (Fig. 2C [resolution 128 × 128] and Fig. 2D [resolution 512 × 512]).
The previously generated 3-D polygonal data and the bone shapes obtained by manipulating the DICOM data in UNITY were precisely positioned in identical spatial coordinates to increase the performance and convenience of the pre-procedural simulation (Fig. 3A and B). A virtual needle was then created to simulate the procedure. The needle was designed to have a shape and size similar to that of the actual needle (Fig. 3C), and colliders were placed in the handle and the needle shaft (Fig. 3D). When the virtual X-ray made contact with these colliders, a value of 2,000 HU was added to the final image so that the needle is expressed as a metallic material on the virtual X-ray image (Fig. 3E and F).
The needle positioning was programmed to move to the target point by holding the approximate position with a mouse and fine-tuning it using keyboard input. In addition, the virtual camera and C-arm were programmed to move and rotate in detail using keyboard input. When the angle of the C-arm was adjusted, the degrees of tilting and rotation were displayed as angles. By another keyboard input, a virtual X-ray could be generated when the desired position of the needle has been obtained in virtual space. Through this simulation, the 3-D positional relationship between the needle and the patient’s anatomy according to the generated X-ray image could be explored in detail.
An 84-year-old woman visited the hospital with a complaint of chronic back pain that radiated to the lateral side of her left leg. Lumbar magnetic resonance imaging (MRI) revealed severe deformity and left foraminal stenosis at the L5/S1 level, which could be responsible for the pain (Fig. 4A). Therefore, transforaminal epidural injection was attempted at the left L5/S1 intervertebral foramen. At the first trial, the authors failed to place the needle in the desired position despite several attempts because her L5/S1 intervertebral foramen was severely narrowed due to deformities and osteophytes. Therefore, a pre-procedural virtual simulation was planned to determine the placement of the needle in the correct position. The simulation was performed using the virtual 3-D data of the patient. After positioning the needle at the optimal position in the anterior epidural space (Fig. 4B), the virtual C-arm was adjusted to the angle at which the needle was viewed as a point (Fig. 4C). Further, several other views were simulated, including the anteroposterior view (Fig. 4D). The spatial relationship between the needle and the spinal anatomy of the patient could be compared in the virtual space. In addition, a 2-D X-ray image could be simulated, which should be obtained in advance for a successful procedure by moving the needle forward while keeping the needle visible as a point. When the patient revisited the hospital, a similar image was obtained using the angle obtained through the simulation, and matched the relative position of the vertebral body and the articular processes by finely adjusting the angle and distance of the actual C-arm, as seen in Fig. 4C. The authors successfully positioned the needle as a pinpoint (Fig. 4E). The needle location was further confirmed through the antero-posterior view (Fig. 4F), and it was found that the contrast medium had appropriately spread into the anterior epidural space.
A 77-year-old man visited the hospital complaining of right pelvic pain due to lung cancer and pelvic bone metastasis. Cancer metastases was observed in the right L5 vertebra and sacral ala of the patient, along with bone marrow edema in the sacroiliac joint in his MRI (Fig. 5A). To ease the severe pain reported by the patient, a superior hypogastric plexus block was planned, which is a challenging procedure since the needle must travel a long distance to the ventral lumbosacral area by avoiding the osteophytes of the transverse process and vertebral body. He had cancer metastasis at the L5 vertebra and sacrum, and subsequent degenerative changes with osteophytes made it more difficult to get the needle to the target. In addition, the transdiscal approach could not be attempted due to the narrow intervertebral disc height of the L5/S1.
We carefully planned a pre-procedural virtual simulation using virtual 3-D data (Fig. 5B). The postero-lateral approach was used and the appropriate angle and direction of the needle that reached the lower L5 anterolateral surface of the vertebral body from the entry point starting near the L4 vertebral body was investigated. The relative position of the C-arm was recorded when the needle was viewed as a point (Fig. 5C). The antero-posterior view of the virtual X-ray was saved, as shown in Fig. 5D. During the next visit of the patient, the C-arm angle was adjusted to obtain the same view as shown in Fig. 5C and inserted the actual 25-gauge needle as a point without difficulty (Fig. 5E). The location of the needle was confirmed with the contrast medium in the anteroposterior view as shown in Fig. 5F.
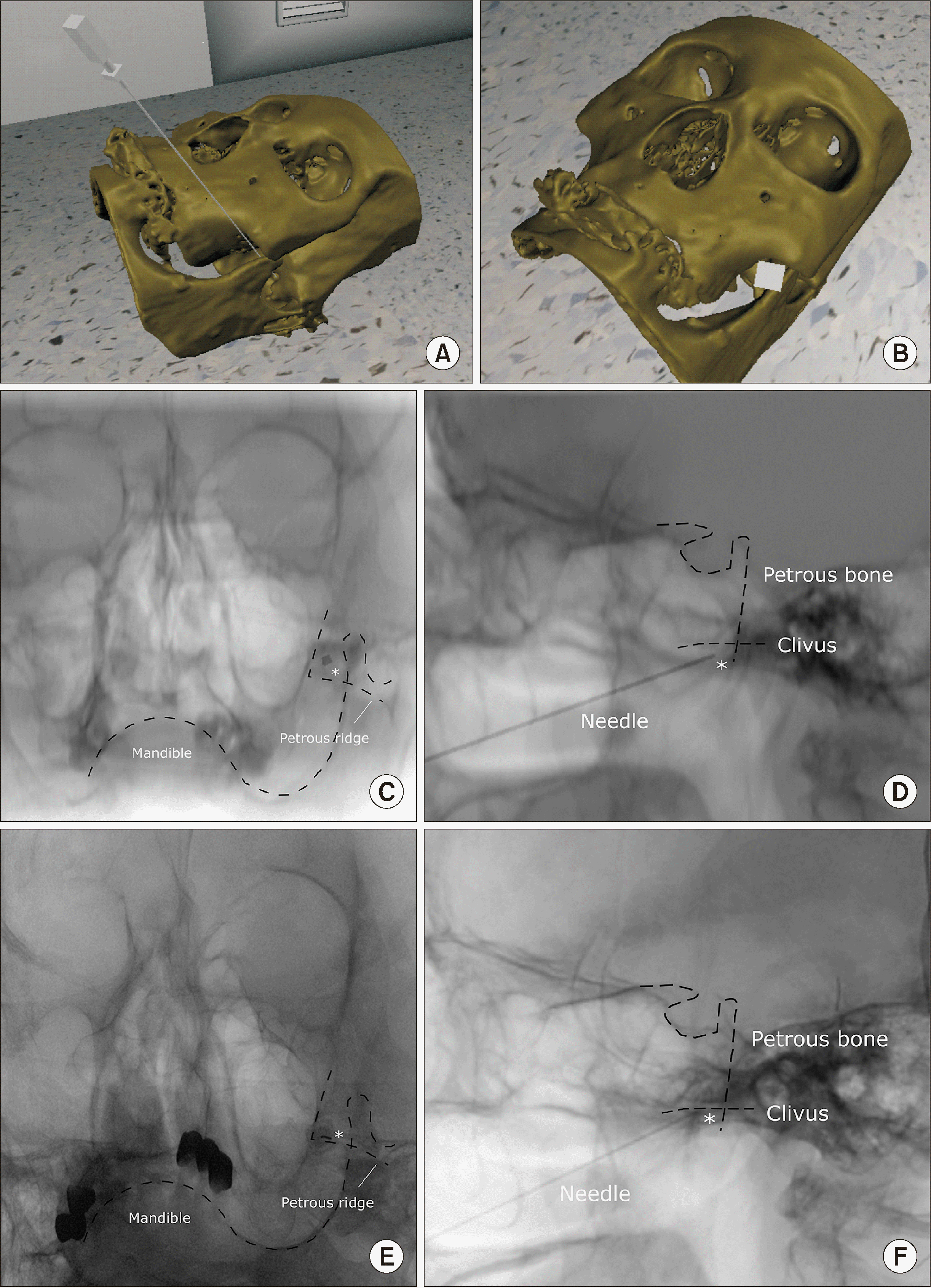
A 69-year-old woman visited the hospital complaining of pain in the left cheek that had begun suddenly and felt like stabbing and electric shock (as described by the patient). A pain a score of 8–9 was assigned by the patient on the numerical rating scale. The pain lasted for 1–2 minutes and became worse with cold stimuli and on eating. A diagnosis of trigeminal neuralgia was established and a Gasserian ganglion block was planned using an anterior approach to relieve the symptoms of the patient. However, in this procedure it is often challenging to identify the target point (i.e., the foramen ovale) even with aid of fluoroscopic guidance, and due to her degenerative changes near the target, there was a fear of injury to important nearby structures. A pre-procedural virtual simulation was attempted to understand the complex structures of the skull. The skull bone was embodied in a virtual simulation using head CT. After accurately positioning the needle in the foramen ovale (Fig. 6A and B), the virtual C-arm was adjusted to view the needles as a point and saved the lateral view (Fig. 6C and D).
Subsequently, when the patient visited the hospital, the foramen ovale was approached as implemented in the virtual simulation, and the procedure was performed easily without complications (Fig. 6E and F).
In this study, a pre-procedural virtual simulation was developed for patients who were expected to experience difficulty in interventional procedures. The simulation was successfully applied to challenging cases with severe deformities.
Computer-integrated virtual simulations have been attempted to aid interventional procedures over the past decade. In terms of preoperative planning, there have been attempts to use preoperative virtual/augmented technologies when surgeries are complex due to the deformation of blood vessels, presence of tumors adjacent to nerves [2,3], or for challenging procedures such as pelvic reconstruction [6]. However, these preoperative simulations have only focused on understanding the challenging anatomy of the patients.
However, for successful pain procedures, it is sometimes insufficient to identify only the deformed anatomy, because even for experienced clinicians who know how the deformed anatomy feels, it is still difficult to accurately position the needle into the target nerve or space with the guidance of 2-D X-ray. Therefore, it is necessary to understand how the needle is expressed on a 2-D X-ray when it is accurately positioned in the deformed anatomy. To the best of the authors’ knowledge, this is the first study that simulates a surgical tool represented by virtual X-rays simultaneously with visualization of the patient’s anatomy that led to the success of an actual procedure.
In this study, a virtual X-ray simulation model was developed. Though there were other studies that created virtual X-rays using CT data and a movable C-arm [7–10], there were few simulation systems that directly access patient data and perform simulations that meet the needs of clinicians. It was essential to program the C-arm with the movement of surgical tools as in the actual operating room so that the clinician could use it directly and conveniently. A DICOM file was used that contains information such as the spatial location, size, and direction of each datum [11,12].
In addition, unlike simulations made by other commercial companies [10,13], this study presents a virtual simulation made by a medical doctor who has been treating patients with pain for a long time. Since all development, including coding, was done by the clinician, the authors believe that the appropriate simulations were developed and can be used for the required treatment area as needed. There are an increasing number of patients with deformed spines who need to undergo challenging procedures. Through virtual simulation, it will be possible to apply treatment tailored for each patient and help understand various complex techniques by reflecting the needs of the clinicians.
The abovementioned three procedures (i.e., transforaminal epidural injection, superior hypogastric plexus block, and Gasserian ganglion block) are commonly performed but can be challenging. With age, many patients develop severe spinal deformities such as lumbar degenerative spondylolisthesis, facet joint hypertrophy, thickening of the ligamentum flavum, spinal curvature, and neural foraminal osteophytes. It is challenging to identify these deformities using 2-plane fluoroscopy [14], especially for transforaminal epidural injection where the needle must enter the anterior epidural space through the intervertebral foramen. In this case, it is advantageous to introduce the needle by avoiding the bony structure as much as possible and keeping it as far as possible from the possible nerve path and blood vessels during the procedure.
The superior hypogastric plexus block has various technical and safety issues. As in this study, there are anatomical impediments (e.g., iliac crest and transverse process of L5) in the path of needle entry for the posterolateral approach, making it challenging to perform. Due to these problems, transdiscal, transvascular, transvaginal, and anterior approaches have been devised [15,16]; however, caution is required because there is a possibility of injury to the intervertebral disc, common iliac artery, bladder, ureter, and bowel [17–19]. Therefore, an accurate skin entry and needle entry angle on an individualized basis are important, and a careful approach is required. By locating the needle in advance to avoid the iliac crest and transverse process with pre-procedural virtual simulation, the insertion point and entry path of the needle can be identified in advance, thus reducing complications during the procedure.
The Gasserian ganglion block is also very complex. This block mainly uses an anterior approach called the “Härtel Method,” which uses bony landmarks to insert a needle through the foramen ovale into Meckel’s cave [20]. It is fundamentally challenging to confirm anatomical landmarks through X-ray images due to the complex structure of the skull [21], and there could be catastrophic complications [22–24]. Various approaches, including CT-guided methods, have been proposed to overcome these difficulties [21,25]. However, none of these methods demonstrate how the needle is expressed on the X-ray when the needle is accurately positioned at the time of the actual procedure. Through the simulation, the exact entry point in the skin can be set, and the exact needle angle can be secured without expensive equipment and increased radiation exposure.
Our study has several limitations. Although the virtual lumbar anatomy was constructed from CT data generally obtained in the supine position, the procedures are performed in the prone position. There might be a difference in spine curvature between the virtual X-ray and the actual X-ray, although this difference could be compensated to some extent through a supporting pillow below the patient’s abdomen in a prone position. In addition, because the virtual X-ray was generated using mainly the coronal and sagittal views of the CT, the quality of the image obtained when the angle was deep was problematic. Also, the lumbar CT images used were not of high resolution. Three prerequisites such as high image resolution, accurate image data registration technique, and software tools for subdividing structures should be well-formed to build a desirable virtual simulation [2]. However, the authors believe that if a clinician makes a simulation themselves and requests the radiology department to obtain the necessary CT data, more efficient simulation will be possible.
The program introduced in this study is currently designed to simulate the procedure of inserting a single needle in advance. However, since it is a simulator that matches the surgical tool and anatomical structure through virtual X-ray generation, it also can be applied in various medical fields, such as other intraoperative guide equipment or artificial intelligence learning. It is hoped that this will contribute to the further development of various medical fields through more applications in the future.
In conclusion, the authors presented creation of a pre-procedure virtual simulation and it was successfully applied on patients who were expected to undergo difficult procedures.
Notes
REFERENCES
1. Bernardo A. 2017; Virtual reality and simulation in neurosurgical training. World Neurosurg. 106:1015–29. DOI: 10.1016/j.wneu.2017.06.140. PMID: 28985656. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85030221433&origin=inward.

2. Kockro RA, Serra L, Tseng-Tsai Y, Chan C, Yih-Yian S, Gim-Guan C, et al. 2000; Planning and simulation of neurosurgery in a virtual reality environment. Neurosurgery. 46:118–35. DOI: 10.1093/neurosurgery/46.1.118. PMID: 10626943. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=18544401605&origin=inward.

3. Mutter D, Dallemagne B, Bailey C, Soler L, Marescaux J. 2009; 3D virtual reality and selective vascular control for laparoscopic left hepatic lobectomy. Surg Endosc. 23:432–5. DOI: 10.1007/s00464-008-9931-y. PMID: 18443871. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=65249137590&origin=inward.

4. Chou R, Gordon DB, de Leon-Casasola OA, Rosenberg JM, Bickler S, Brennan T, et al. 2016; Management of postoperative pain: a clinical practice guideline from the American Pain Society, the American Society of Regional Anesthesia and Pain Medicine, and the American Society of Anesthesiologists' Committee on Regional Anesthesia, Executive Committee, and Administrative Council. J Pain. 17:131–57. Erratum in: J Pain 2016; 17: 508-10. DOI: 10.1016/j.jpain.2015.12.008. PMID: 26827847. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84959275025&origin=inward.

5. Ehlke M, Ramm H, Lamecker H, Hege HC, Zachow S. 2013; Fast generation of virtual X-ray images for reconstruction of 3D anatomy. IEEE Trans Vis Comput Graph. 19:2673–82. DOI: 10.1109/TVCG.2013.159. PMID: 24051834. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84886715427&origin=inward.

6. Shen F, Chen B, Guo Q, Qi Y, Shen Y. 2013; Augmented reality patient-specific reconstruction plate design for pelvic and acetabular fracture surgery. Int J Comput Assist Radiol Surg. 8:169–79. DOI: 10.1007/s11548-012-0775-5. PMID: 22752350. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84878888400&origin=inward.

7. Allen DR. 2020. Simulation approaches to X-ray C-arm-based interventions [master's thesis]. The University of Western Ontario;London (ON): https://ir.lib.uwo.ca/etd/7231/.
8. De Silva T, Punnoose J, Uneri A, Mahesh M, Goerres J, Jacobson M, et al. 2018; Virtual fluoroscopy for intraoperative C-arm positioning and radiation dose reduction. J Med Imaging (Bellingham). 5:015005. DOI: 10.1117/1.JMI.5.1.015005. PMID: 29487882. PMCID: PMC5812884. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85042444462&origin=inward.

9. De Silva T, Punnoose J, Uneri A, Goerres J, Jacobson M, Ketcha MD, et al. 2017; C-arm positioning using virtual fluoroscopy for image-guided surgery. Proc SPIE Int Soc Opt Eng. 10135:101352K. DOI: 10.1117/12.2256028. PMID: 28572694. PMCID: PMC5449120. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85020447848&origin=inward.

10. Touchette M, Newell R, Anglin C, Guy P, Lefaivre K, Amlani M, et al. 2021; The effect of artificial X-rays on C-arm positioning performance in a simulated orthopaedic surgical setting. Int J Comput Assist Radiol Surg. 16:11–22. DOI: 10.1007/s11548-020-02280-2. PMID: 33146849. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85094898276&origin=inward.

11. Muschelli J. 2019; Recommendations for processing head CT data. Front Neuroinform. 13:61. DOI: 10.3389/fninf.2019.00061. PMID: 31551745. PMCID: PMC6738271. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85075822608&origin=inward.

12. Sattler B, Lee JA, Lonsdale M, Coche E. 2010; PET/CT (and CT) instrumentation, image reconstruction and data transfer for radiotherapy planning. Radiother Oncol. 96:288–97. DOI: 10.1016/j.radonc.2010.07.009. PMID: 20709416. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=77956188071&origin=inward.

13. Våpenstad C, Buzink SN. 2013; Procedural virtual reality simulation in minimally invasive surgery. Surg Endosc. 27:364–77. DOI: 10.1007/s00464-012-2503-1. PMID: 22956001. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84875827513&origin=inward.

14. Clarençon F, Law-Ye B, Bienvenot P, Cormier É, Chiras J. 2016; The degenerative spine. Magn Reson Imaging Clin N Am. 24:495–513. DOI: 10.1016/j.mric.2016.04.008. PMID: 27417397. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84979031451&origin=inward.

15. Choi JW, Kim WH, Lee CJ, Sim WS, Park S, Chae HB. 2018; The optimal approach for a superior hypogastric plexus block. Pain Pract. 18:314–21. DOI: 10.1111/papr.12603. PMID: 28520297. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85021780736&origin=inward.

16. Lee YW, Yoon DM, Lee GM, Han ST, Park HJ. 2000; Trans-intervertebral disc approach of superior hypogastric plexus block for pelvic cancer pain a retrospective study. J Korean Pain Soc. 13:202–7.
17. Bosscher H. 2001; Blockade of the superior hypogastric plexus block for visceral pelvic pain. Pain Pract. 1:162–70. DOI: 10.1046/j.1533-2500.2001.01017.x. PMID: 17129292.

18. Urits I, Schwartz R, Herman J, Berger AA, Lee D, Lee C, et al. 2021; A comprehensive update of the superior hypogastric block for the management of chronic pelvic pain. Curr Pain Headache Rep. 25:13. DOI: 10.1007/s11916-020-00933-0. PMID: 33630172. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85101922424&origin=inward.

19. Rocha A, Plancarte R, Nataren RGR, Carrera IHS, Pacheco V, Hernandez-Porras BC. 2020; Effectiveness of superior hypogastric plexus neurolysis for pelvic cancer pain. Pain Physician. 23:203–8. DOI: 10.36076/ppj.2020/23/203. PMID: 32214302.
20. Nurmikko TJ, Eldridge PR. 2001; Trigeminal neuralgia--pathophysiology, diagnosis and current treatment. Br J Anaesth. 87:117–32. DOI: 10.1093/bja/87.1.117. PMID: 11460800. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=0034942939&origin=inward.

21. Matys T, Ali T, Zaccagna F, Barone DG, Kirollos RW, Massoud TF. 2019; Ossification of the pterygoalar and pterygospinous ligaments: a computed tomography analysis of infratemporal fossa anatomical variants relevant to percutaneous trigeminal rhizotomy. J Neurosurg. 132:1942–51. DOI: 10.3171/2019.2.JNS182709. PMID: 31075780. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85085993638&origin=inward.

22. Brown JA. 1997; Direct carotid cavernous fistula after trigeminal balloon microcompression gangliolysis: case report. Neurosurgery. 40:886. DOI: 10.1097/00006123-199704000-00060. PMID: 9092871. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=0030933275&origin=inward.

23. James EA, Kibbler CC, Gillespie SH. 1995; Meningitis due to oral streptococci following percutaneous glycerol rhizotomy of the trigeminal ganglion. J Infect. 31:55–7. DOI: 10.1016/S0163-4453(95)91457-9. PMID: 8522835. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=0029148995&origin=inward.

24. Rath GP, Dash HH, Bithal PK, Goyal V. 2009; Intracranial hemorrhage after percutaneous radiofrequency trigeminal rhizotomy. Pain Pract. 9:82–4. DOI: 10.1111/j.1533-2500.2008.00246.x. PMID: 19019055. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=58249126256&origin=inward.

25. Bohnstedt BN, Tubbs RS, Cohen-Gadol AA. 2012; The use of intraoperative navigation for percutaneous procedures at the skull base including a difficult-to-access foramen ovale. Neurosurgery. 70(2 Suppl Operative):177–80. DOI: 10.1227/NEU.0b013e3182309448. PMID: 21822157.

Fig. 1
Process of acquiring the patient’s image in a virtual space. (A) The computed tomographic image is obtained and positioned in the virtual space by using the values of image orientation, image position, and pixel spacing values. (B) When the Hounsfield unit value of 200–400 is only visualized, a bone-like three-dimensional image could be acquired.

Fig. 2
Virtual X-ray generation. Virtual X-rays are generated from the generator to the inside of the four corners of the detector, and the spatial coordinates are divided by the desired resolutions (A, 5 × 5) and (B, 10 × 10). The movement of the virtual C-arm is programmed to mimic the movements of the actual C-arm, with adjustments made using a keyboard. After calculating the point on the computed tomography slice where each X-ray collision occurred, the corresponding Hounsfield unit values are obtained. The degree of absorption of the X-rays is calculated and subsequently converted to the pixel value (C, resolution 128 × 128 and D, resolution 512 × 512).

Fig. 3
Creation of a procedure needle expressed on a virtual X-ray. (A, B) Patient’s Digital Imaging and Communications in Medicine and polygonal data are positioned in the same virtual space to manipulate the simulation conveniently. (C) Creation of the virtual needle similar to the actual needle. (D) Colliders are set in the handle and the needle shaft. (E, F) When the virtual X-ray collides with the collider of the needle, a value of 2,000 is added so that it can be expressed as a metallic material in the virtual X-ray.
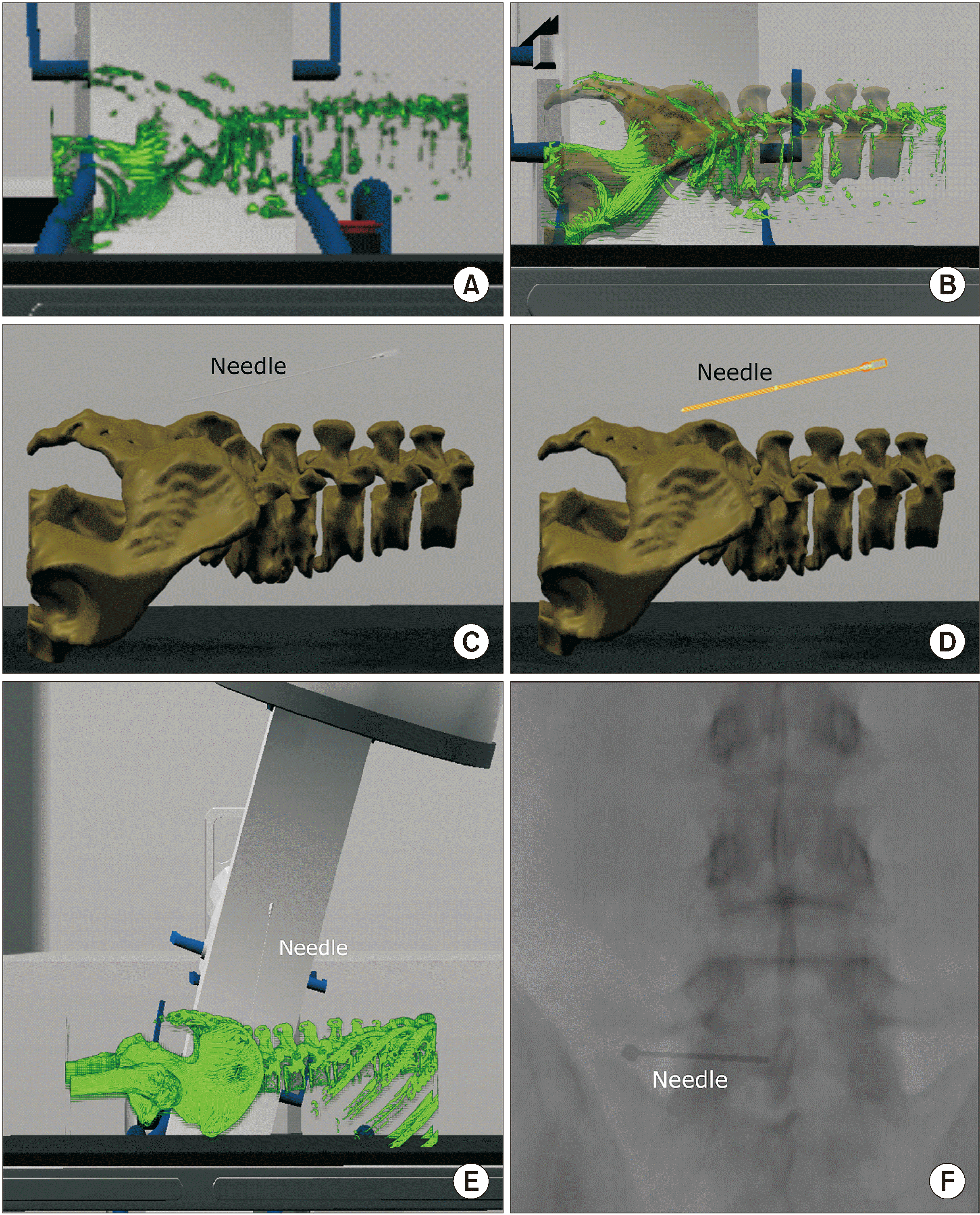
Fig. 4
Application of virtual simulation before performing transforaminal epidural injection in a patient with severe spinal deformity. (A) Axial plane of T2-weighted magnetic resonance imaging (MRI). Lumbar MRI of the patient revealed left sided L5 nerve root irritation due to severe spinal deformity seemed to be the cause of pain. (B) Three-dimensional virtual procedural planning. The virtual needle is placed in the most appropriate position. (C) Oblique view of the virtual X-ray image is saved where the needle presents as a pinpoint. (D) Anteroposterior view of the simulated virtual X-ray. (E) The patient’s oblique X-ray image during the procedure. (F) The patient’s X-ray image in the anteroposterior view during the procedure. White asterisk: needle, TP: transverse process, SAP: superior articular process, Dotted lines: contour of anatomical structures.
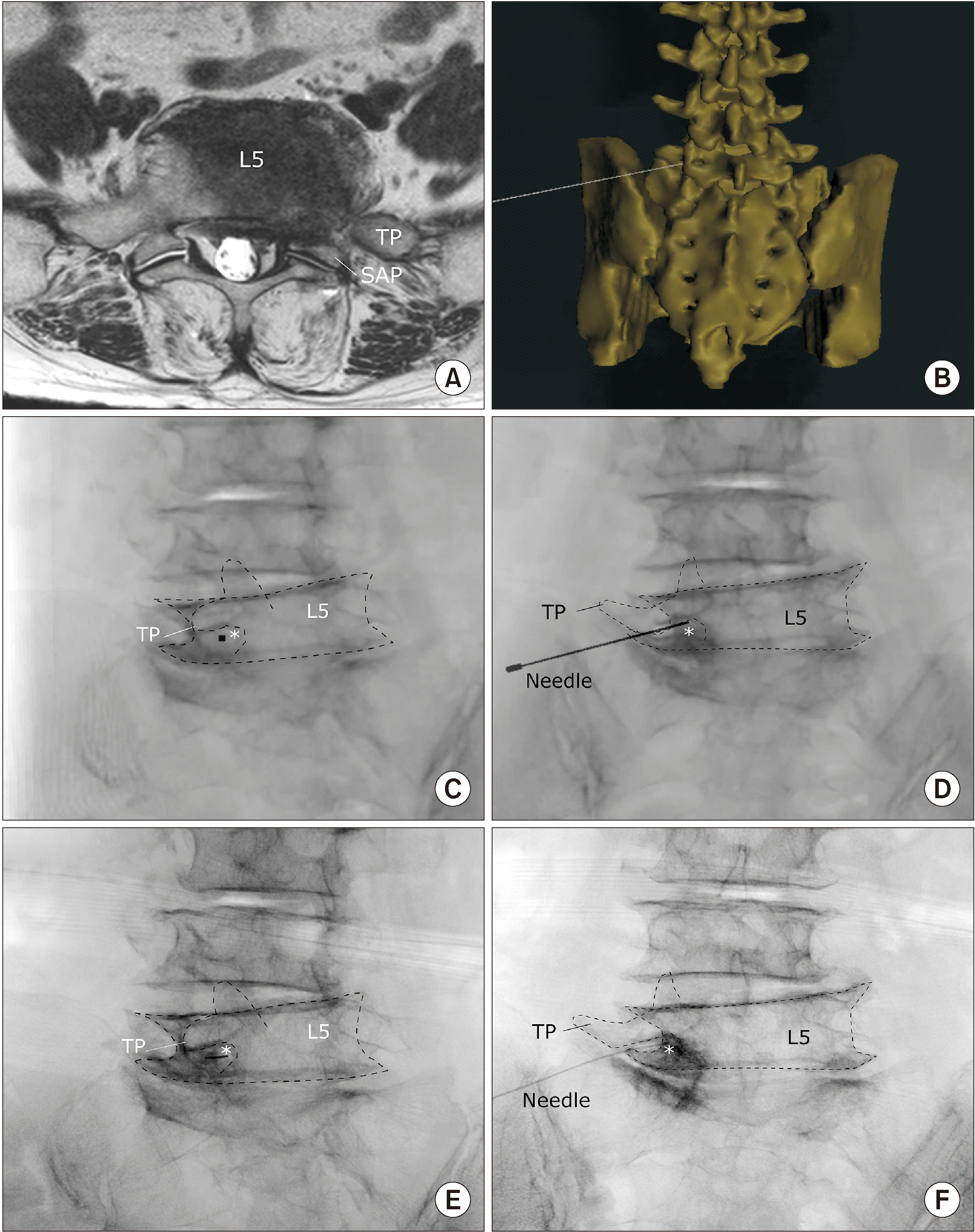
Fig. 5
Application of virtual simulation in performing the superior hypogastric plexus block. (A) Coronal view of the patient’s magnetic resonance image. Metastasis to the right sided L5 vertebra and sacral ala seems to cause the pain. (B) Three-dimensional virtual procedural planning. The virtual needle is placed at the most appropriate position. (C) Oblique view of the virtual X-ray image is saved where the needle presents as a pinpoint. (D) Anteroposterior view of the simulated virtual X-ray. (E) The patient’s oblique X-ray image during the procedure. (F) The patient’s X-ray image in the anteroposterior view during the procedure. White asterisk: needle, TP: transverse process, SAP: superior articular process, Dotted lines: contour of anatomical structures.
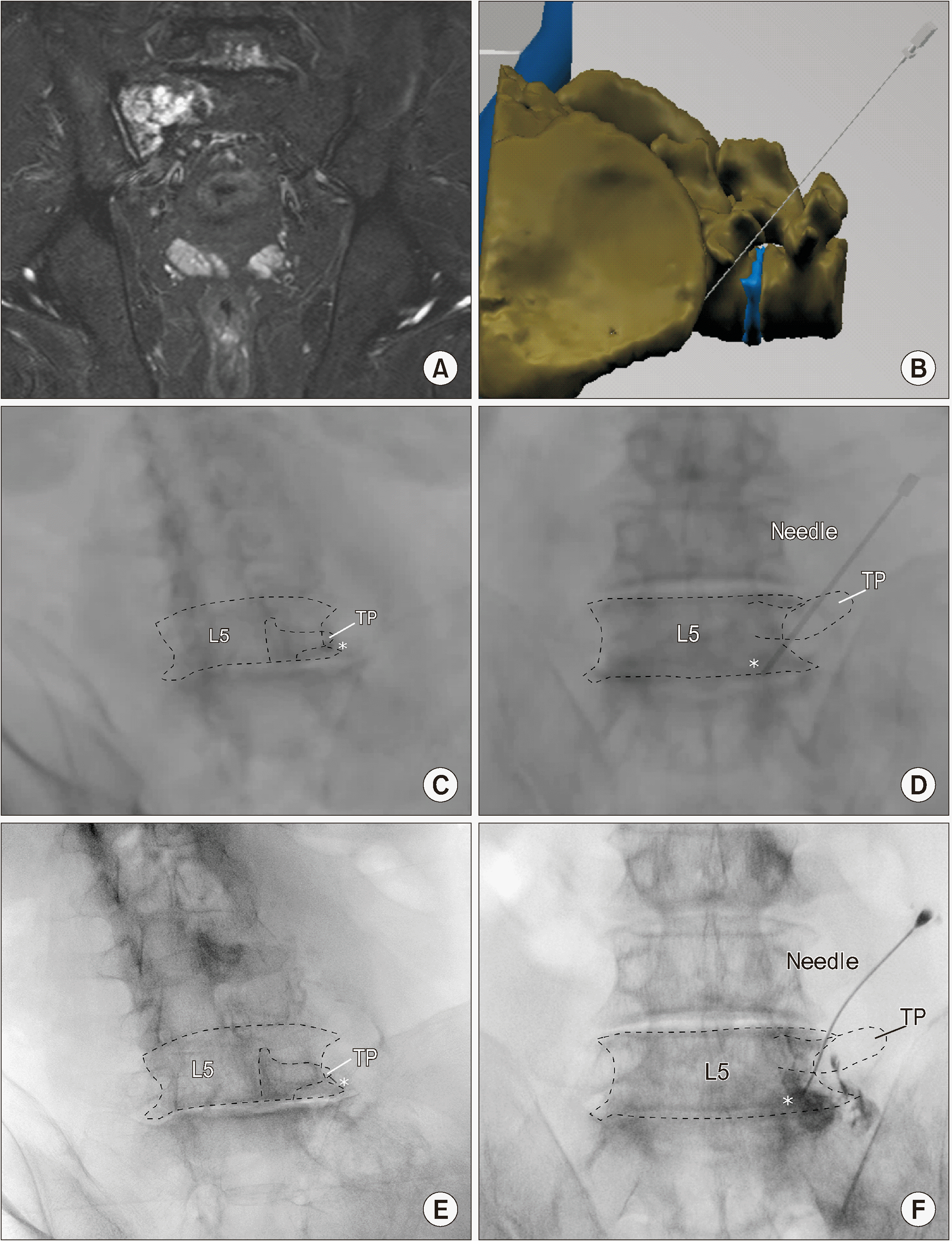
Fig. 6
Application of virtual simulation in performing the Gasserian ganglion block. (A, B) Three-dimensional virtual procedural planning. The virtual needle is placed at the most appropriate position with reference to the right foramen ovale. (C) Caudal tilted anteroposterior view of the virtual X-ray image is saved where the needle presented as a pinpoint. (D) Lateral view of the simulated virtual X-ray. (E) The patient’s X-ray image in the caudal tilted anteroposterior view during the procedure. (F) The patient’s X-ray image in the lateral view during the procedure. White asterisk: needle, Dotted lines: contour of anatomical structures.




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download