Abstract
It has been reported that overconsumption of caffeine during pregnancy leads to a deleterious effect within the nervous tissues during embryonic development. In this study, we further extrapolated the effect of caffeine in the developing retinas, which is known to be one of the most sensitive tissues in chick embryos. Morphological changes of retinal thickness and organization of neuroretinal epithelium were monitored using three gene markers, Atoh7, FoxN4, and Lim1. Upon treating with a single dose of caffeine (15 µmol at embryonic day 1 [E1]), relative thicknesses of developing retinas (particularly of E7 and E9) were significantly altered. Among the three genes studied, the expression pattern of Atoh7 was notably altered while those of FoxN4, and Lim1 mRNA showed only a slight change in these developing retinas. Quantitative polymerase chain reaction results supported the most notable changes of Atoh7 but not FoxN4, and Lim1 gene in the developing retinas, particularly at E7. The effect of caffeine towards other organs during development should be extrapolated and the awareness of its intensive consumption should be raised.
Retinogenesis is a complex event that is involved in cellular organization in the definitive period which has been well studied in vertebrates, including chick and mouse models. Based on the periods of differentiation, retina progenitor cells (RPCs) have been divided into 2 subgroups, early-born group (retinal ganglion cell [RGC], horizontal cell [HC], and amacrine cell [AC]) and late-born group (cone photoreceptor, bipolar cell, rod photoreceptor, and Müller cell). The molecular controls of retinogenesis have been extensively documented, and many well-characterized molecules have been anticipated as the universal biomarkers for the key steps of retinal development [1, 2]. Our focus in this study is on the early-born population and their markers since they are the targets for many teratogens.
One of the common teratogens is caffeine, a white crystalline xanthine alkaloid, which exerts both positive and negative effects on the central nervous system (CNS). Its beneficial effects on recovering signs and symptoms of the CNS disorders have been demonstrated in the cases of Parkinson’s disease, Alzheimer’s disease, and glaucoma [3-5]. Alternatively, caffeine consumption in pregnant women has raised concern as a result of its increasing evidence in its interruption of organogenesis during the early development of embryos. The adverse effect of caffeine has been reported in many highly suspectable organs including heart, skeleton, gonad, neural tissue; brain, spinal cord, and retina [6-12]. Due to the structural homology of caffein and adenosine nucleoside, caffeine is known to interact with adenosine receptors (ARs) which antagonistically modulates an inhibition of phosphodiesterase enzyme and the release of intracellular calcium ions through the G-protein and cyclic adenosine monophosphate (cAMP)-independent signaling cascade [3, 13]. Effect of caffeine during retinal development has also been reported to be involved in 2 types of ARs, AR1 and AR2A, expressed in the inner and outer plexiform layers (IPL and OPL) from E10 until E17 of chick retinas [11]. The exposure to caffeine subsequently leads to down-regulation of both AR1 and AR2A in the developing retinas [14]. Apart from ARs, there are also a number of caffeine-responsive receptors such as GABAA-, ryanodine-, glycine-, gustatory-, and dopaminergic- receptors that are found in Drosophila [15-18]. Interestingly, dopaminergic receptors (DRs) are also detected in the early chick developing retinas, D1A and D1B depending on developmental ages [19]. These DRs also share a common pathway with ARs leading to an increase in the level of cAMP [20]. It has been reported that at early development of E9 (without the AR expression), caffeine affects retinal morphology causing a reduction of retinal thickness [20]. We thus further extrapolated here the possible selective susceptibility of caffeine towards any given types of neuroretinal cells in the early-born population of developing retina.
During early retinal development, the orchestrated molecular controls to synthesize cellular organization in developing retinas has been well documented [1, 2]. In this study, we focused on the 3 marker genes which are spaciotemporal expressed in the 3 population of neuroepithelial cells, Atoh7 (for RGC), FoxN4 (for AC and HC), and Lim1 (for HC). Firstly, Atoh7 (atonal bHLH transcription factor 7) is a transcription factor in retinogenesis which plays an essential role in RGC differentiation [20]. In mouse embryos, the expression of Atoh7 is initially detected at E12 in RPC for specification and is gradually declined during the postnatal age [21-23]. Secondly, FoxN4 (Forkhead box N4) is a transcription factor in neurogenesis that exists in humans, mouse, chicken, xenopus, and zebrafish [24]. The expression of FoxN4 is involved in the genesis of early-born retinal cells including AC and HC which is a further required specifier for cell specialization [25-27]. Lastly, we focused on the specific regulator of FoxN4-lineage, Lim1 (Lim homeobox 1) which is involved in the development of female reproductive tract and head. In order to differentiate into HC, Lim1 is required as a downstream of FoxN4 to promote HC differentiation [27, 28]. In this study, we injected a single dose of caffeine at a concentration of 15 µmol/egg into the E1 embryo and observed the structural and genetic changes at E3, E5, E7, and E9. Alterations of the universal retinal genetic markers (Atoh7, FoxN4, and Lim1) are evident by in situ hybridization and real-time polymerase chain reaction (PCR) and the possible disturbance of caffeine towards differentiation and migration of the three populations of developing retino-neuronal cells including RGC, HC, and AC is discussed.
Fertilized chick eggs, Gallus gallus were obtained from Department of Animal Science, Faculty of Agriculture, Kasetsart University, Thailand. Handling of these embryos followed the guidelines of the Animal Care Committee, Faculty of Science, Bangkok (protocol no. MUSC61-029-431). The eggs were incubated at 38°C with a relative humidity of 60% on the rotatory plate. To administer the caffeine, a small window was created on the eggshell at the blunt end (air chamber). Thereafter, caffeine solution at a concentration of 15 µmol/egg was injected into a yolk sac of E1 fertilized eggs and the window was sealed with parafilm. Control eggs were injected with 0.9% normal saline solution with the same volume. All administered and control embryos were then cultured in the 38°C incubator.
Administered chick embryos at E3–E9 were harvested by cracking the shell and dissected free from yolk matter. The eyes of embryos were carefully removed from the heads and fixed in 4% paraformaldehyde in 0.12 M phosphate buffer for either 6 hours (E3) or overnight (E5–E9) at 4°C. For paraffin embedding, the tissue was further dehydrated in the increasing percentage of ethanol, infiltrated in xylene, and finally embedded in paraffin. The 5 µm-thick sections were cut and routinely processed for hematoxylin and eosin staining. The images were acquired by a Nikon E600 light microscope (Nikon Instruments Inc., NY, USA) and Panoramic Digital slide Scanner (3D-Histech; 3DHISTECH Kft., Budapest, Hungary). They were further processed and analyzed by Adobe Photoshop CS6 software (Peachpit Press, Berkeley, CA, USA) to measure the retinal thickness. The measurement of retinal thickness was conducted at the retinal area which is opposite the mid-center point of the posterior lens surface. Statistical analysis was performed by paired t-test using a GraphPad Prism 9 software (Graphpad Inc., San Diego, CA, USA). A P-value <0.05 was considered to be statistically significant.
For cryo-sectioning, they were transferred into 10% sucrose in phosphate buffer overnight at 4°C and further submersed and embedded in gelatin-sucrose (14% gelatin, 0.1% agarose, 10% sucrose solution) and kept at –80°C. Sections were cut by a Leica RM2235 rotatory cryo-microtome at –20°C (Leica, Buffalo Grove, IL, USA) at a thickness of 25 µm and adhered onto Matsunami platinum microscopic slides (Matsunami Glass, Bellingham, WA, USA). All images were acquired, processed, and analyzed as mentioned above.
Riboprobes of 400 to 500 nucleotides were designed and cloned in pBluescript II SK+ plasmids (General Biosystem, Durham, NC, USA) for constructing Atoh7, FoxN4, and Lim1 probes. Antisense probes were synthesized by in vitro transcription using T3 RNA polymerase (Roche, Basel, Switzerland) and DIG-RNA labeling mix (Roche). They were precipitated with cold ethanol and 4M LiCl2 overnight at –20°C and recovered in diethylpyrocarbonate-treated distilled water.
In situ hybridization was applied according to previously described protocol [29]. Cryo-sections were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS), washed in PBS and treated with 20 μg/ml proteinase K (Roche) in PBS. They were further permeabilized with 1% Triton-X 100 in PBS and incubated with pre-hybridization buffer (50% formamide, 5× saline-sodium citrate [SSC], 5× Denhardt, 250 µg/ml yeast tRNA, 500 µg/ml Salmon sperm DNA) at room temperature for 2 hours. Tissues were then incubated with RNA probe in hybridization buffer (1:200) and sealed with a cover glass at 72°C for overnight in a moist chamber. After 24-hour incubation, the cover glass was removed and the samples were immersed in 0.2×SSC at 72°C and in buffer I (1 M Tris-HCl, pH 7.6, and 5 M NaCl). They were further transferred into 10% normal goat serum (Abcam, Cambridge, UK) in buffer I (room temperature, 2 hours). Then, the sections were incubated with 1:5,000 goat anti-digoxygenin conjugated with alkaline phosphatase (Roche) in 1% blocking solution (4°C, overnight) in a moist chamber. The samples were washed in buffer I and submersed in buffer III (1 M Tris-HCl, pH 9.5 and 5 M NaCl, 1 M MgCl2, 0.1% Tween 20; Vivantis Technologies Sdn. Bhd., Selangor, Malaysia). The enzymatic product was developed by 100 ng/ml nitro blue tetrazolium (NBT), 50 mg/ml 5-bromo-4-chloro-3-indolyl-phosphate (BCIP) in buffer III, rinsed in PBS, and mounted with 1:1 glycerol and PBS (v/v) prior to topping with a cover glass.
Caffeine treated- and controlled- chick retinas were isolated, and the retinal pigment epithelium and lens were carefully removed. Isolated retinas were then subjected to a total RNA extraction using a Trizol reagent (Life Technology, Carlsbad, CA, USA) and Direct-zol RNA Mini Prep (ZYMO Research, Tustin, CA, USA) and resuspended in a DNase/RNase-free water. Approximately, 1 µg of RNA was reversely transcribed by an iScript Reverse Transcription Supermix for RT-qPCR (Bio-Rad, Hercules, CA, USA). Complementary DNA (cDNA) was amplified using a Luna Universal qPCR master mix (BioLabs, Ipswich, MA, USA) containing 10 µM primers (Table 1) and 0.5 µg of cDNA, in a total volume of 10 µl. Thermal cycle conditions were 95°C for 1 minute, 53.9°C for 30 seconds and 72°C for 30 seconds run at 44 cycles. The data were presented as mean±standard error of the mean. The differences between groups were analyzed by one-way ANOVA followed by the Bonferroni post-test by using a Graphpad Prism 9 software (Graphpad Inc.). A P-value <0.05 was considered to be statistically significant.
The histological structures and their morphometric parameters (thickness and cellular organization) between caffeine-treated and control chick embryos (E3–E9) were compared and shown in Table 2. The measured thicknesses of retinas in both groups were 47.49±4.62 (n=8) vs. 62.18±10.53 for E3 (n=5); 70.85±9.78 (n=7) vs. 80.38±6.50 for E5 (n=6); 203.27±18.21 (n=5) vs. 140.09±9.24 for E7 (n=6); and 254.81±27.52 (n=9) vs. 215.30±23.83 for E9 (n=7). Interestingly, significant differences in the epithelial thickness were noted in E7 (P<0.01, double asterisk) and E9 (P<0.05, asterisk) groups. The cellular organization appeared rather similar in both groups (Fig. 1). In E3 to E7, stacking of cells forming neuroblastic layer (NBL) towards presumptive ganglion cell layer (pGCL) in the innermost layer was well observed in both control (Fig. 1A–C, I–K) and caffeine-treated retinas (Fig. 1E–G, M–O). At E9 where all definitive epithelial layers including outer nuclear layer (ONL), OPL, inner nuclear layer (INL), IPL, and GCL were clearly identified in the normal developing retina (Fig. 1L), they were also well-identified in caffeine-treated animals (Fig. 1P). Particularly, the IPL (located between GCL and INL, representing the first delamination process of GCL between the two nuclear layers) could still be clearly visible in both groups (asterisk in Fig. 1L, P).
Among the three markers studied, expression of Atoh7 mRNA appeared to be significantly altered in all aged embryos upon being treated with caffeine. The expression of Atoh7 was detected at the outermost layer of the developing retina (the site where the RPC-proliferative cells were localized) in the E3-control embryos (Fig. 2A). Similarly, its expression was also detectable at the outermost layer of the E3-caffeine treated embryos (Fig. 2E). At E5 of the control group, the expression of Atoh7 was additionally detected in the innermost layer or pGCL where RGC cell population was localized (asterisk in Fig. 2B) apart from that of the outermost layer. In contrast, Atoh7 remained slightly detected in the outermost layer while it was non-detectable in the pGCL layer of the caffeine-treated E5 group (Fig. 2F). At E7 and E9 of the control animals, similar expression pattern of Atoh7 was observed as with E5-control where the staining was found in GCL, but not in outermost NBL. In the same vein, the expression of Atoh7 was also observed in the GCL and outermost layer of NBL in the E7 and ONL in the E9 caffeine-treated embryos (Fig. 2G, H).
Two other markers that were studied herein were FoxN4 and Lim1 which have been used to differentiate AC and HC lineages. Localization of these two mRNA seemed to be minimally altered after the animals were subjected to caffeine treatment. As shown in Fig. 2I–X, the expressions of FoxN4 and Lim1 mRNA were still silent in E3 control embryos both in control and caffeine injected embryos. At E5, FoxN4 was expressed in the innermost layer of pGCL and Lim1 was found to be more specific to the inner layer of pGCL in the control animals (Fig. 2J, R). The staining pattern of both FoxN4 and Lim1 appeared to be scattered within this NBL layer in the caffeine-treated animals (Fig. 2N, V). In the control group of E7, localization of FoxN4 was relatively different from that of Lim1, in which FoxN4 was not detected in the retina while Lim1 was observed at the vicinity of pGCL, an anticipated localization of the migratory HCs in the control group (Fig. 2S) as well as in ONL and GCL at E9 (Fig. 2T). In comparison to the caffeine-treated group, the expression of FoxN4 was detected as faint staining in the middle layer of INL (Fig. 2P), while the expression of Lim1 had no notable change on the animals treated with caffeine (Fig. 2X).
We also quantitated the expression levels of the three marker genes collected from E7 and E9 embryos (n=3 each) using real-time PCR analysis (Fig. 3). We had to confess that in our handling method, the retinas of E3 and E5 were too small to be collected and it was difficult obtaining enough RNA, therefore, only data of E7 and E9 were presented here. The results of qPCR well agreed with those of in situ hybridization, and Atoh7 gene expression was mostly affected by caffeine treatment, while the other two markers showed minimal gene alterations. Specifically, the level of Atoh7 gene expression was reduced by 85% (0.15±0.01 relative fold change) and 57% (0.43±0.03 relative fold change) in E7 and E9, respectively (P<0.05). The relative fold changes of FoxN4 level at E7 and E9 were not significantly altered, with the values of 0.99±0.13 (or 1% reduction) for E7 and 0.64±0.13 (or 36% decrease) for E9. A similar trend of change was also observed for Lim1 where the gene levels were 0.84±0.21 (16% reduction) for E7 and 0.99±0.21 (1% decrease) for E9.
During retinogenesis, many teratogens are known to cause a change of the retinal formation and maturation. Since caffeine is the most consumable bioactive alkaloid, its effects could potentiate both beneficial and adverse outcomes. It has been reported in human that caffeine can pass freely through the placenta and blood-brain barrier and it can potentially affect development by lowering body length and weight of newborn [30-33]. Similar effect has been proven in the chick embryo model in which exposure to caffeine at concentrations of 5, 10, and 15 µmol/egg at the early stage-embryos (the highest dose) directly disrupts the expression and function of Pax6 which consequently leads to an asymmetric microphthalmia with a reduction in size of lens and retinal thickness [33]. These findings can be attributed to increase in reactive oxygen species which can be inhibited by the pre-administration of vitamin C to counter the teratogenic effect of caffeine [20]. In our study, chick embryos were also employed to test the teratogenic effect of a single-dose of caffeine at 15 µmol/egg in which the alteration of gene expression was notable. Most significantly, the alteration in Atoh7 expressing cells that is normally found in RGC population of E5 developing embryos was reciprocally switched off after caffeine treatment. Alternatively, there were very slight changes (both morphologically and quantitatively) of the other two markers, FoxN4 and Lim1 (Figs. 2, 3). Thus far, there is no direct correlation between the action of caffeine that is modulated on any specific type of receptor(s) that affect the expression levels of specific genes related to retinal development, particularly, the 3 genes that were selected herein. The example mentioned above where the expression of Pax6 is downregulated upon caffeine administration would require further extrapolation on caffeine-receptor interaction leading to its downstream molecular consequences, such as Atoh7.
One of the caffeine-responsive receptors whose effect on many developing tissues have been most well studied is the AR which may be, due to the structural homolog between caffeine and adenosine molecules. In this regard, the effect of caffeine on developing retinas may not be out of expectation as many previous studies have reported that ARs were localized in the retinal of chicken, rabbit, mice, rats, monkeys, and human [15, 34-36]. However, it has also been reported that the expression of both AR receptors in the developing chick retinas starts to be observed at E12 to E15, the timing which does not well correspond with the expression of the three marker genes studied herein. Other receptors expressed during the early embryonic period and have been reported to have interfered with caffeine should thus be more considerable. As earlier mentioned, DR1A is expressed as early as E7 embryos, its interruption by caffeine antagonistic binding would interfere with developing genes or transcription factors expressed in this given period [19]. The best example earlier mentioned is the expression of Pax6 transcriptional factor which is also known to be interrupted by caffeine treatment [11]. In fact, Pax6 (as well as Sox2) are the upstream regulators of Atoh7 that are expressed within RPC population once the stem cells are committed to be GCL cell lineage [37]. Therefore, interference of Atoh7 (particularly at GCL; Fig. 2) and the reduced retinal thickness at E7 and E9 would thus well agree with this previous finding [33]. However, the non-responsiveness of FoxN4 and Lim1 mRNA (both of which are the other downstream cascade of Pax6 that further differentiate into AC and HC lineages) towards caffeine administration would present a further interesting issue of whether the signal transductions of these lineages are upregulated by a different type of caffeine-sensitive receptors as well as regulated by different molecular cascades apart from Pax6. Regardless of its molecular mechanism, one possible conclusion that could be drawn from our study is that early exposure to caffeine during early development leads to a structural change of the retina in which the thickness of neuroepithelium and cellular organization is significantly altered (Fig. 2). It is more likely that caffeine would cause a delay or alteration of the organization pattern of the specific population in the neuroepithelial cells (in our case, Atoh7-expressing RGCs). More extensive investigation is thus required to explore this complex involvement of caffeine in retinogenesis. Nevertheless, healthcare issues should be raised to sensitize consumers about the risk of caffeine that could potentiate retinal hazards and thus newborns’ visual ability.
Acknowledgements
This study was supported by a grant from the Development and Promotion of Science and Technology Talents Project (DPST), Thailand. We also thank the Center of Nanoimaging (CNI) and the Central Instrument Facility (CIF), Faculty of Science, Mahidol University, for providing instrumental support throughout this work.
Notes
Author Contributions
Conceptualization: TL, WW, S Asuvapongpatana. Data acquisition: TL, AB, S Ahi. Data analysis or interpretation: TL, AB, NC. Drafting of the manuscript: TL, S Asuvapongpatana. Critical revision of the manuscript: WW, S Asuvapongpatana. Approval of the final version of the manuscript: all authors.
References
1. Reinhardt R, Centanin L, Tavhelidse T, Inoue D, Wittbrodt B, Concordet JP, Martinez-Morales JR, Wittbrodt J. 2015; Sox2, Tlx, Gli3, and Her9 converge on Rx2 to define retinal stem cells in vivo. EMBO J. 34:1572–88. DOI: 10.15252/embj.201490706. PMID: 25908840. PMCID: PMC4474531.

2. Xiang M. 2013; Intrinsic control of mammalian retinogenesis. Cell Mol Life Sci. 70:2519–32. DOI: 10.1007/s00018-012-1183-2. PMID: 23064704. PMCID: PMC3566347.

3. Ribeiro JA, Sebastião AM. 2010; Caffeine and adenosine. J Alzheimers Dis. 20(Suppl 1):S3–15. DOI: 10.3233/JAD-2010-1379. PMID: 20164566.

4. Espinosa J, Rocha A, Nunes F, Costa MS, Schein V, Kazlauckas V, Kalinine E, Souza DO, Cunha RA, Porciúncula LO. 2013; Caffeine consumption prevents memory impairment, neuronal damage, and adenosine A2A receptors upregulation in the hippocampus of a rat model of sporadic dementia. J Alzheimers Dis. 34:509–18. DOI: 10.3233/JAD-111982. PMID: 23241554.

5. Madeira MH, Elvas F, Boia R, Gonçalves FQ, Cunha RA, Ambrósio AF, Santiago AR. 2015; Adenosine A2AR blockade prevents neuroinflammation-induced death of retinal ganglion cells caused by elevated pressure. J Neuroinflammation. 12:115. DOI: 10.1186/s12974-015-0333-5. PMID: 26054642. PMCID: PMC4465153.

6. Rana N, Moond M, Marthi A, Bapatla S, Sarvepalli T, Chatti K, Challa AK. 2010; Caffeine-induced effects on heart rate in zebrafish embryos and possible mechanisms of action: an effective system for experiments in chemical biology. Zebrafish. 7:69–81. DOI: 10.1089/zeb.2009.0631. PMID: 20415645.

7. Hawkins JA, Hu N, Clark EB. 1984; Effect of caffeine on cardiovascular function in the stage 24 chick embryo. Dev Pharmacol Ther. 7:334–43. DOI: 10.1159/000457182. PMID: 6478987.

8. Ma ZL, Qin Y, Wang G, Li XD, He RR, Chuai M, Kurihara H, Yang X. 2012; Exploring the caffeine-induced teratogenicity on neurodevelopment using early chick embryo. PLoS One. 7:e34278. DOI: 10.1371/journal.pone.0034278. PMID: 22470550. PMCID: PMC3314624. PMID: 7d0e0aa9839f42c7bccf14f64deea72e.

9. Park M, Choi Y, Choi H, Yim JY, Roh J. 2015; High doses of caffeine during the peripubertal period in the rat impair the growth and function of the testis. Int J Endocrinol. 2015:368475. DOI: 10.1155/2015/368475. PMID: 25983753. PMCID: PMC4423020.

10. Dorostghoal M, Erfani Majd N, Nooraei P. 2012; Maternal caffeine consumption has irreversible effects on reproductive parameters and fertility in male offspring rats. Clin Exp Reprod Med. 39:144–52. DOI: 10.5653/cerm.2012.39.4.144. PMID: 23346524. PMCID: PMC3548072.

11. Brito R, Pereira-Figueiredo D, Socodato R, Paes-de-Carvalho R, Calaza KC. 2016; Caffeine exposure alters adenosine system and neurochemical markers during retinal development. J Neurochem. 138:557–70. DOI: 10.1111/jnc.13683. PMID: 27221759.

12. Sheth S, Brito R, Mukherjea D, Rybak LP, Ramkumar V. 2014; Adenosine receptors: expression, function and regulation. Int J Mol Sci. 15:2024–52. DOI: 10.3390/ijms15022024. PMID: 24477263. PMCID: PMC3958836.

13. de Carvalho RP, Braas KM, Adler R, Snyder SH. 1992; Developmental regulation of adenosine A1 receptors, uptake sites and endogenous adenosine in the chick retina. Brain Res Dev Brain Res. 70:87–95. DOI: 10.1016/0165-3806(92)90106-7. PMID: 1473280.

14. Mustard JA. 2014; The buzz on caffeine in invertebrates: effects on behavior and molecular mechanisms. Cell Mol Life Sci. 71:1375–82. DOI: 10.1007/s00018-013-1497-8. PMID: 24162934. PMCID: PMC3961528.

15. Lee Y, Moon SJ, Montell C. 2009; Multiple gustatory receptors required for the caffeine response in Drosophila. Proc Natl Acad Sci U S A. 106:4495–500. DOI: 10.1073/pnas.0811744106. PMID: 19246397. PMCID: PMC2657413.
16. Moon SJ, Köttgen M, Jiao Y, Xu H, Montell C. 2006; A taste receptor required for the caffeine response in vivo. Curr Biol. 16:1812–7. DOI: 10.1016/j.cub.2006.07.024. PMID: 16979558.
17. Nall AH, Shakhmantsir I, Cichewicz K, Birman S, Hirsh J, Sehgal A. 2016; Caffeine promotes wakefulness via dopamine signaling in Drosophila. Sci Rep. 6:20938. DOI: 10.1038/srep20938. PMID: 26868675. PMCID: PMC4751479.

18. Soares HC, de Melo Reis RA, De Mello FG, Ventura AL, Kurtenbach E. 2000; Differential expression of D(1A) and D(1B) dopamine receptor mRNAs in the developing avian retina. J Neurochem. 75:1071–5. DOI: 10.1046/j.1471-4159.2000.0751071.x. PMID: 10936188.

19. Neve KA, Seamans JK, Trantham-Davidson H. 2004; Dopamine receptor signaling. J Recept Signal Transduct Res. 24:165–205. DOI: 10.1081/RRS-200029981. PMID: 15521361.

20. Ma ZL, Wang G, Cheng X, Chuai M, Kurihara H, Lee KK, Yang X. 2014; Excess caffeine exposure impairs eye development during chick embryogenesis. J Cell Mol Med. 18:1134–43. DOI: 10.1111/jcmm.12260. PMID: 24636305. PMCID: PMC4508153.

21. Brown NL, Patel S, Brzezinski J, Glaser T. 2001; Math5 is required for retinal ganglion cell and optic nerve formation. Development. 128:2497–508. DOI: 10.1242/dev.128.13.2497. PMID: 11493566. PMCID: PMC1480839.
22. Yang Z, Ding K, Pan L, Deng M, Gan L. 2003; Math5 determines the competence state of retinal ganglion cell progenitors. Dev Biol. 264:240–54. DOI: 10.1016/j.ydbio.2003.08.005. PMID: 14623245.

23. Le TT, Wroblewski E, Patel S, Riesenberg AN, Brown NL. 2006; Math5 is required for both early retinal neuron differentiation and cell cycle progression. Dev Biol. 295:764–78. DOI: 10.1016/j.ydbio.2006.03.055. PMID: 16690048.

24. Boije H, Edqvist PH, Hallböök F. 2008; Temporal and spatial expression of transcription factors FoxN4, Ptf1a, Prox1, Isl1 and Lim1 mRNA in the developing chick retina. Gene Expr Patterns. 8:117–23. DOI: 10.1016/j.modgep.2007.09.004. PMID: 18006384.

25. Edqvist PH. 2006. Neuronal development in the embryonic retina: focus on the characterization, generation and development of horizontal cell subtypes [PhD dissertation]. Acta Universitatis Upsaliensis;Uppsala:
26. Gouge A, Holt J, Hardy AP, Sowden JC, Smith HK. 2001; Foxn4--a new member of the forkhead gene family is expressed in the retina. Mech Dev. 107:203–6. DOI: 10.1016/S0925-4773(01)00465-8. PMID: 11520680.
27. Li S, Mo Z, Yang X, Price SM, Shen MM, Xiang M. 2004; Foxn4 controls the genesis of amacrine and horizontal cells by retinal progenitors. Neuron. 43:795–807. DOI: 10.1016/j.neuron.2004.08.041. PMID: 15363391.

28. Dyer MA, Livesey FJ, Cepko CL, Oliver G. 2003; Prox1 function controls progenitor cell proliferation and horizontal cell genesis in the mammalian retina. Nat Genet. 34:53–8. DOI: 10.1038/ng1144. PMID: 12692551.

29. Marillat V, Sabatier C, Failli V, Matsunaga E, Sotelo C, Tessier-Lavigne M, Chédotal A. 2004; The slit receptor Rig-1/Robo3 controls midline crossing by hindbrain precerebellar neurons and axons. Neuron. 43:69–79. DOI: 10.1016/j.neuron.2004.06.018. PMID: 15233918.

30. Klebanoff MA, Levine RJ, Clemens JD, Wilkins DG. 2002; Maternal serum caffeine metabolites and small-for-gestational age birth. Am J Epidemiol. 155:32–7. DOI: 10.1093/aje/155.1.32. PMID: 11772782.

31. Bracken MB, Triche EW, Belanger K, Hellenbrand K, Leaderer BP. 2003; Association of maternal caffeine consumption with decrements in fetal growth. Am J Epidemiol. 157:456–66. DOI: 10.1093/aje/kwf220. PMID: 12615610.

32. Bakker R, Steegers EA, Obradov A, Raat H, Hofman A, Jaddoe VW. 2010; Maternal caffeine intake from coffee and tea, fetal growth, and the risks of adverse birth outcomes: the generation R study. Am J Clin Nutr. 91:1691–8. DOI: 10.3945/ajcn.2009.28792. PMID: 20427730.

33. Momoi N, Tinney JP, Liu LJ, Elshershari H, Hoffmann PJ, Ralphe JC, Keller BB, Tobita K. 2008; Modest maternal caffeine exposure affects developing embryonic cardiovascular function and growth. Am J Physiol Heart Circ Physiol. 294:H2248–56. DOI: 10.1152/ajpheart.91469.2007. PMID: 18359892.

34. Blazynski C, Perez MT. 1991; Adenosine in vertebrate retina: localization, receptor characterization, and function. Cell Mol Neurobiol. 11:463–84. DOI: 10.1007/BF00734810. PMID: 1683815.

35. Braas KM, Zarbin MA, Snyder SH. 1987; Endogenous adenosine and adenosine receptors localized to ganglion cells of the retina. Proc Natl Acad Sci U S A. 84:3906–10. DOI: 10.1073/pnas.84.11.3906. PMID: 3473489. PMCID: PMC304985.

36. Kvanta A, Seregard S, Sejersen S, Kull B, Fredholm BB. 1997; Localization of adenosine receptor messenger RNAs in the rat eye. Exp Eye Res. 65:595–602. DOI: 10.1006/exer.1996.0352. PMID: 9367639.

37. Nguyen-Ba-Charvet KT, Rebsam A. 2020; Neurogenesis and specification of retinal ganglion cells. Int J Mol Sci. 21:451. DOI: 10.3390/ijms21020451. PMID: 31936811. PMCID: PMC7014133.

Fig. 1
Histology of developing retinas of control and caffeine-treated embryos at the stages of E3–E9. The paraffin sections were stained with hematoxylin and eosin and the images were acquired at low magnification (bar 200 µm, A–H) and high magnification (bar 100 µm, I–P) to demonstrate cellular organization in retinas. NBL, neuroblastic layer; pGCL, presumptive ganglion cell layer; GCL, ganglion cell layer; IPL, inner plexiform layer; INL, inner nuclear layer; OPL, outer plexiform layer; ONL, outer nuclear layer. *Delaminated IPL.

Fig. 2
Localization of the three retinal marker genes in developing retinas of the control and caffeinetreated embryos. Note the apparent changes in the staining pattern and intensity of Atoh7 gene in E7 and E9 retinas of control (A–D) and caffeine-treated animals (E–H), while those of FoxN4 (I–P), and Lim1 (Q–X) are minimally changed. Bars in E3 column=100 μm; the E5, E7, E9 column=50 μm. NBL, neuroblastic layer; pGCL, presumptive ganglion cell layer; GCL, ganglion cell layer; IPL, inner plexiform layer; INL, inner nuclear layer; OPL, outer plexiform layer; ONL, outer nuclear layer.
Fig. 3
Quantitative analysis of the three retinal marker genes in embryos treated with a single dose of caffeine compared with the control embryos. Total of RNA of the whole eyes were collected from E7 and E9 embryos and the expression levels of Atoh7 (A), FoxN4 (B), and Lim1 (C) were quantitated by real-time polymerase chain reaction (PCR). The data were calculated from 2–δδCq quantitative PCR values of caffeine-treated and control embryos and expressed as mean±standard error of log2 fold changes of the triplicated.
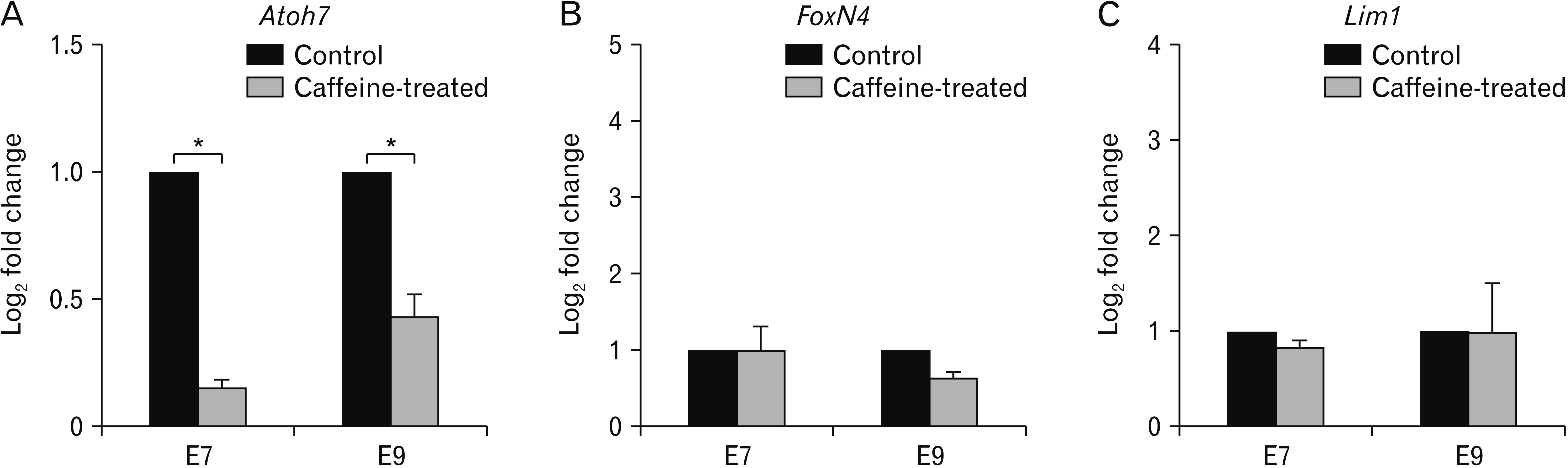
Table 1
Primers that are used for real-time polymerase chain reaction
| Gene | Accession number | Primer sequences | Product size (bp) |
|---|---|---|---|
| Atoh7_Forward | NM_204668.1 | GAGAATGGATTAACCTTCACTGTGAAC | 99 |
| Atoh7_Reverse | GCTGTGCATAAGGATCACTGTCTG | ||
| FoxN4_Forward | NM_001083359.1 | AGCACCTGGAGCAATGATTC | 131 |
| FoxN4_Reverse | CTGTAAACTAGGAGAGGGC | ||
| Lim1_Forward | NM_205413.1 | GCGGGCTGTGCCCAGGGCAT | 140 |
| Lim1_Reverse | TTTTCGTCTATGATATAGAG | ||
| Gapdh_Forward | NM_204305.1 | ATGATCCCTTCATCGATCTG | 106 |
| Gapdh_Reverse | ATCACAAGTTTCCCGTTCTC |
Table 2
Thickness and cellular organization in control and caffeine-treated retinas
| Embryonic stage | Average of retinal thickness (µm) | Number of cell layers (layer) | |||
|---|---|---|---|---|---|
| Control | Caffeine-treated | Control | Caffeine-treated | ||
| E3 | 47.49±4.62 | 62.18±10.53 | 1 (NBL) | 1 (NBL) | |
| E5 | 70.85±9.78 | 80.38±6.50 | 1 (NBL) | 1 (NBL) | |
| E7 | 203.27±18.21 | 140.09±9.24** | 2 (pGCL, NBL) | 2 (pGCL, NBL) | |
| E9 | 254.81±27.52 | 215.30±23.83* | 5 (GCL, IPL, INL, OPL, ONL) | 5 (GCL, IPL, INL, OPL, ONL) | |




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download