This article has been
cited by other articles in ScienceCentral.
Abstract
Background
The pathophysiology underlying primary adult immune thrombocytopenic purpura (ITP) has not yet been identified. However, many mechanisms affect the immune system, causing defective tolerance to self-platelets and megakaryocytes. Cluster of differentiation 40 (CD40) contributes to both humoral and cell-mediated immune responses.
Methods
This case‒control study was conducted to detect rs4810485G>T and rs1883832C>T polymorphisms of CD40 in Egyptian patients with persistent/chronic ITP to clarify their possible association with chronic disease evolution. This study included 50 patients with persistent/chronic ITP and 50 healthy controls. Genotyping was performed using the polymerase chain reaction‒restriction fragment length polymorphism technique.
Results
Genotyping of rs1883832 and rs4810485 revealed no statistically significant differences between the two groups. However, combined gene polymorphism genotyping showed a statistically significant difference between the two groups (P<0.01).
Conclusion
Our results indicate a strong association between the combined polymorphism of both genes and susceptibility to developing ITP among adult Egyptian patients. Targeting this pathway using novel therapeutic approaches is promising.
Keywords: CD40, rs1883832, rs4810485, Polymorphism, Chronic ITP
INTRODUCTION
Immune thrombocytopenic purpura (ITP) is an autoimmune disease of unknown etiology. Both genetic and environmental factors are thought to play a role in disease development. Several genes involved in immune system regulation, including cytokines, FcγR, cytotoxic T-lymphocyte-associated protein-4 (CTLA-4), human leukocyte antigen genes, and some infectious agents such as hepatitis C virus (HCV), human immunodeficiency virus, and
Helicobacter pylori (
H. pylori), have been associated with increased susceptibility to ITP in several studies [
1].
Several abnormalities involving the cellular mechanisms of immune modulation have also been described. T-cell dysregulation and T-cell-related cytokine abnormalities, such as T-helper 1/T-helper 2 (Th1/Th2) imbalance, decreased number and dysfunction of regulatory T cells, increased T-lymphocyte-mediated cytotoxicity, and elevated Th17 cells, have been suggested to play a crucial role in the development of ITP [
2].
Cluster of differentiation 40 (CD40), a member of the tumor necrosis family of transmembrane glycoproteins, was found to be rapidly and transiently expressed on the recently activated CD4
+ T-cell surface and is a potent T-cell costimulatory molecule that stimulates autoreactive T cells expressing CD154 (CD40L). CD154 is also expressed by T follicular helper (TFH) cells, localized in the germinal centers of lymphoid organs, which in turn stimulates proliferation, differentiation, and antibody production by B cells. Splenic TFH cells are expanded in ITP and participate in autoreactive B cell activation and antiplatelet antibody production. Megakaryocytes and platelets also express CD154 and can directly activate B cells to produce antiplatelet antibodies. Thus, the CD40/CD154 (CD40L) axis acts as a costimulatory signal at diverse levels [
3]. CD40 is also expressed in monocytes, dendritic cells (DCs), mast cells, basophils, B cells, natural killer cells, and macrophages, highlighting the important role of the CD40 pathway in cellular biology [
4]. Interactions between CD40 and platelet-associated CD154 (CD40L) induce B-cell immunoglobulin (Ig) production, monocyte activation, and DC differentiation. Several polymorphisms in the gene encoding CD40 have been identified, and the relationship between these genetic polymorphisms and risk of developing several autoimmune and inflammatory diseases, such as Graves’ disease and rheumatoid arthritis, has been reported [
5].
A CD40 ligand on the surface of effector T-cells binds to CD40 on the surface of B-cells; however, it does not contain a “death domain”. This binding helps drive the resting B cell into the cell cycle and is essential for B-cell responses to thymus-dependent antigens [
6]. Moreover, CD40 signaling in B cells promotes germinal center formation, Ig isotype switching, Ig somatic hypermutation to increase affinity for antigens, and finally formation of long-lived plasma cells and memory B cells [
7].
CD40L expression is increased in patients with ITP and stimulates the activation of autoreactive B lymphocytes. Blockade of the CD40/CD154 signal is a potential immunomodulatory strategy for T-cell-mediated diseases, and many studies have suggested that blocking this signaling is effective for ITP through selective suppression of autoreactive T and B lymphocytes to platelet antigens [
8].
Moreover, the rs1883832C>T polymorphism of CD40 increases CD40 expression by upregulating the transcription or translation efficiency of the CD40 gene, and the abnormal expression of CD40 increases pro-inflammatory cytokines, causing the development of diseases [
9].
This study was conducted to detect the possible association between two single-nucleotide polymorphisms (SNPs) rs1883832 and rs4810485 in the CD40 gene and susceptibility to chronic adult ITP in adolescent and adult Egyptian populations using a polymerase chain reaction–restriction fragment length polymorphism (PCR-RFLP) assay. The results of this study could be the basis for future immunomodulatory therapies that could help in the treatment of persistent/chronic ITP.
MATERIALS AND METHODS
This study included 50 patients diagnosed with persistent/chronic ITP at the Hematology Outpatient Clinic of El Kasr Al Ainy Hospital, Faculty of Medicine, Cairo University, between April 2016 and December 2017 and 50 age- and sex-matched healthy Egyptian volunteers as the control group. ITP was diagnosed according to the American Society of Hematology [
10]. The control group included healthy subjects with normal platelet counts and no history of blood diseases. The age of the patients ranged from 14 to 60 years, with a mean age of 30.6±11.12 years. The study group included five male patients (10%) and 45 female patients (90%). The age of the patients in the control group ranged from 16 to 53 years, with a mean value of 30.9±9.3 years; seven (14%) of them were male and 43 (86%) were female. Clinical and laboratory data of the study group are shown in
Table 1.
The study protocol was approved by the Ethics Committee of Kasr Al Ainy faculty, Cairo University, and conducted in accordance with the Declaration of Helsinki (ethical principles for medical research involving human subjects).
Adult Egyptian patients diagnosed with persistent/chronic ITP aged 14–60 years were selected to participate, whereas those with secondary causes of ITP, such as HCV, systemic lupus erythematosus (SLE), malignancy, and drug-induced thrombocytopenia were excluded.
All patients enrolled in the study were subjected to full history taking and thorough clinical examination to exclude organomegaly and lymphadenopathy and look for signs of secondary causes of ITP, such as malar rash of SLE or swollen tender joints, together with routine laboratory workup, including complete blood count, and other laboratory investigations to exclude secondary causes of ITP, including antinuclear antibody testing, HCV antibody, HIV antibody, anticardiolipin immunoglobulin G and immunoglobulin M, C3, and C4. Bone marrow aspiration was performed at the time of diagnosis to exclude underlying malignancies and other hematological diseases.
All patients were followed for >1 year to assess their chronicity. Defining the disease state at the time of sampling according to Neunert
et al. [
10], 50 patients under study (100%) were in activity with a platelet count of <100×10
9/L (persistent ITP with relapse following termination of therapy within 3–12 months from diagnosis), and none (0%) of the patients were in remission (platelet count >100×10
9/L either spontaneously or following therapy).
Genotyping of the
CD40 gene was performed using PCR-RFLP to detect two SNPs within the
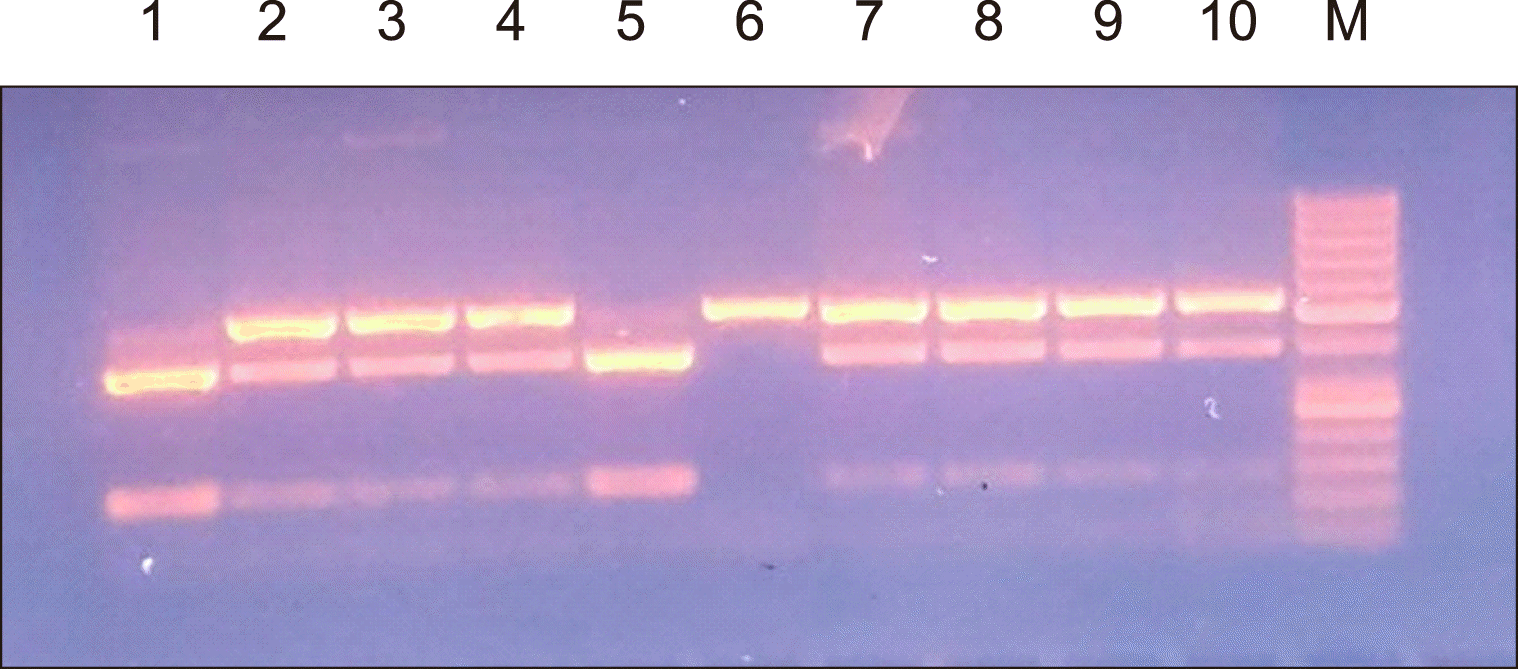
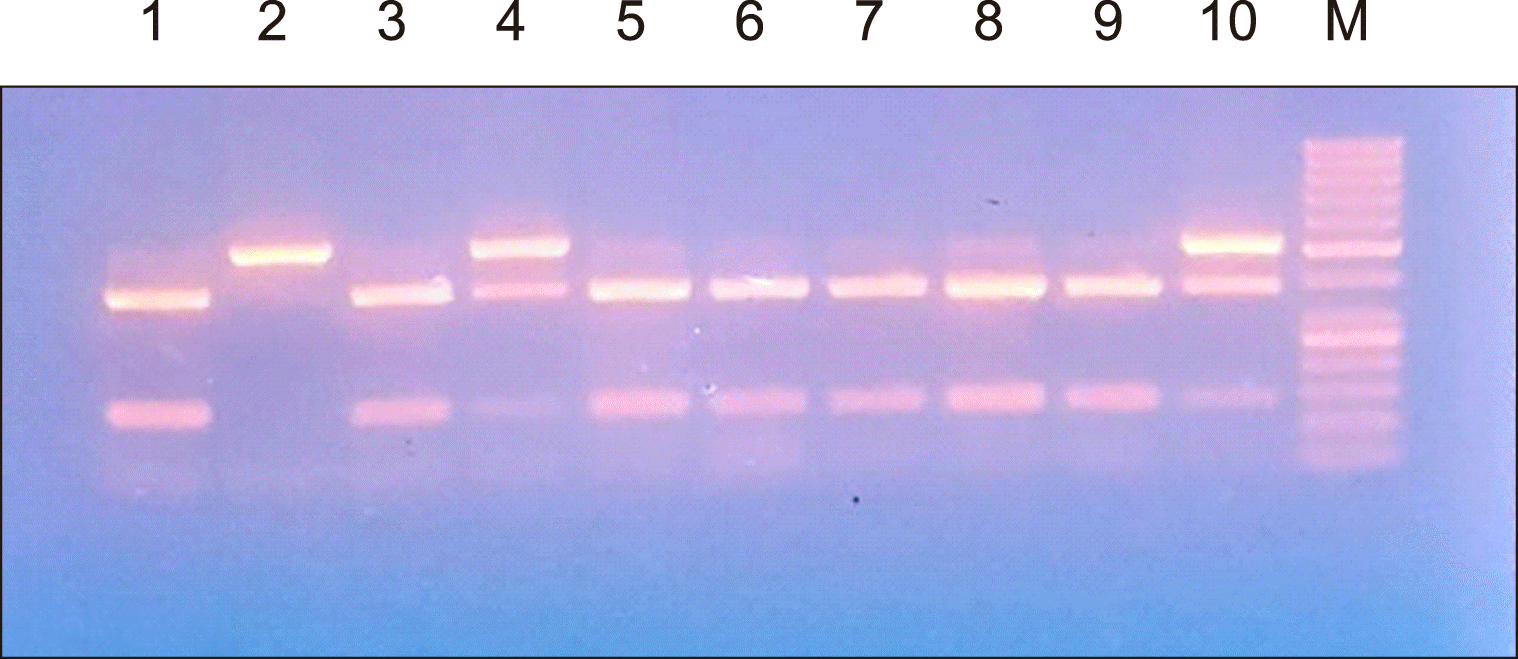
CD40 gene: rs1883832 and rs4810485. The samples were stored at -20°C until analysis. Genomic DNA was extracted using the Quick-gDNA MiniPrep Kit (catalog no: D3024, USA). The primer sequences for the CD40 polymorphisms (rs4810485 and rs1883832) were provided by Biosearch Technologies (California, USA) where the following primers were used: F:5'-CCC CGA TAG GTG GAC CGC GAT TG-3', R:5'-CCC GCC CTC TGA ACC CCC TAC CA-3' for the rs1883832 SNP, and F:5'-TAT TTT TGT AGT TCC TCA TTC TG-3', R:5'-GCC CCC CTT TAC CTC TTT C-3' for the rs4810485 SNP. The extracted DNA was amplified using MyTaq Red Mix (Bioline, Australia). PCR amplification consisted of an initial heating step at 94°C for 2 min, followed by 35 cycles of denaturation (at 94°C for 1 min), annealing (60°C for 1 min), and chain extension (72°C for 1 min), which ended with a final extension (72°C for 7 min), followed by cooling at 4°C. The amplified PCR products of the gene were digested using the restriction enzymes FastDigest NcoI for rs1883832 and FastDigest MspI for rs4810485. The restriction products were subjected to gel electrophoresis in a 2% agarose gel and visualized by staining with ethidium bromide in reference to a molecular weight marker [
11].
For rs1883832 C>T gene polymorphism:
Wild genotype provides a single-band 503 base pair (bp).
Homozygous genotype provides two bands of 130 bp and 373 bp.
Heterozygous genotype provides three bands of 503, 130, and 373 bp.
For rs4810485 G>T polymorphism:
Wild genotype provides one band at 288 bp.
Homozygous genotype provides two bands at 104 bp and 184 bp, respectively.
Heterozygous genotype yields three bands at 288, 104, and 184 bp.
Statistical methods
All collected data were revised to ensure completeness and accuracy. Pre-coded data were entered into a computer using Statistical Package for the Social Sciences (version 21; IBM Corp., Armonk, NY, USA) for the statistical analysis. Data were summarized using mean and standard deviation for quantitative variables and numbers and percentages for qualitative variables. Quantitative variables were compared using the independent t-test for normally distributed variables and nonparametric Mann–Whitney U test for non-normally distributed variables, whereas qualitative variables were compared using the chi-square test. Binary logistic regression was performed to explore significant predictors of ITP. Probability values (P-values) <0.05 were used to denote statistical significance.
RESULTS
In this study, we examined the rs1883832 gene polymorphism where the wild (CC) genotype was encountered in four patients in the study group (8%) and two subjects in the control group (4%). The heterozygous (CT) genotype was found in 24 patients in the study group (48%) and 28 subjects in the control group (56%), whereas the homozygous (TT) genotype was found in 22 patients in the study group (44%) and 20 subjects in the control group (40%). The frequency of the C allele was 32% among patients with ITP and 32% among subjects in the control group, whereas the frequency of the T allele was 68% among patients with ITP and 68% among subjects in the control group.
The comparison between patients with ITP and the control group regarding the rs1883832 (C>T) gene polymorphism as a risk factor for the development of chronic ITP did not show statistically significant difference between the two groups with no liability of patients with the rs1883832 (C>T) gene polymorphism to develop chronic ITP [P>0.05; odds ratios (OR), 0.417, 1.078, and 1.619, respectively; 95% confidence interval (CI), 0.143–1.215, 0.098–11.809, and 0.705–3.716, respectively].
Simultaneously, examination of the rs4810485 gene polymorphism showed that the wild (GG) genotype was found in four patients in the study group (8%) and four patients in the control group (8%); the heterozygous (GT) genotype was found in 24 patients in the study group (48%) and 16 subjects in the control group (32%), whereas the homozygous (TT) genotype was encountered in 22 patients in the study group (44%) and 30 subjects in the control group (60%).
The frequency of the G allele was 32% among patients with ITP and 24% among subjects in the control group, whereas the frequency of the T allele was 68%.
The comparison between patients with ITP and the control group regarding the rs4810485 (G>T) gene polymorphism as a risk factor for the development of chronic ITP revealed no statistically significant difference between the two groups, with no liability of patients with the rs4810485 (G>T) gene polymorphism to develop chronic ITP (P>0.05; OR, 0.733, 1.5, and 0.478, respectively; 95% CI, 0.165–3.257, 0.327–6.882, and 0.201–1.133, respectively).
Patients exhibiting polymorphisms in both genes (combined gene polymorphism) were compared with the control group exhibiting polymorphism of a single gene, and a highly statistically significant difference was observed between the two groups (
P<0.001) (
Table 2).
A comparison between completely responsive, responsive, and non-responsive patients with ITP regarding either the rs1883832 (C>T) or rs4810485 (G>T) genotype and allele did not show a statistically significant difference between the three groups regarding gene polymorphism or allele frequencies (
Table 3).
Agarose gel electrophoresis of the rs1883832 and rs4810485 PCR products are shown in
Fig. 1,
2 respectively.
DISCUSSION
CD40 expression in peripheral blood lymphocytes and platelet-associated CD154 expression are elevated in patients with ITP. CD40L overexpression causes abnormal activation of reactive B lymphocytes; thus, CD40L plays an important role in the pathogenesis of ITP by producing autoantibodies against platelet surface antigens [
8].
Yan
et al. [
12] proposed that CD40 expression is affected, in turn affecting the CD40–CD40L inflammatory signal response pathways, due to
CD40 gene polymorphism rs1883832 C>T, where mutations result in different ITP susceptibility trends in individuals, suggesting that the C allele upregulates the expression of CD40 on B lymphocytes, whereas the
CD40 gene polymorphism rs4810485 and ITP susceptibility are irrelevant in a Liaoning Han population.
In this case–control study, the rs1883832 C>T and rs4810485 G>T genotypes were examined by PCR amplification of the target gene, followed by allele-specific restriction enzyme digestion (RFLP) in 50 adult patients with persistent/chronic ITP and 50 age- and sex-matched healthy Egyptian volunteers as the control group. Genotyping of rs1883832 C>T revealed no statistically significant differences between the patient and control groups (P>0.05). Moreover, rs4810485 G>T genotyping revealed no statistically significant differences between the patient and control groups (P>0.05).
Combined gene polymorphism genotyping revealed that the frequencies of GG-CC, GT-CT, and TT-TT among patients with ITP were 8%, 48%, and 44%, respectively, compared to 0%, 16%, and 28% in the control group, indicating a strong statistically significant difference (P<0.01) that may suggest that the presence of mutant alleles is a risk factor for ITP.
In another Egyptian study, AbdelGhafar
et al. [
13] evaluated the impact of
CD40 gene polymorphisms on the risk of developing ITP in 101 patients with ITP and 97 healthy subjects. The two SNPs of the
CD40 gene (rs1883832 C/T and rs4810485 G/T) were genotyped using TaqMan allele discrimination real-time PCR, and
CD40 (rs1883832) TT genotype carriers had a significantly higher risk of ITP than CC genotype carriers (adjusted OR, 3.792; 95% CI, 1.252–11.49;
P=0.018). The T allele also represented a 1.711-fold increased risk of ITP, which was more evident in men (
P=0.016). No significant difference was observed in the frequency of CD40 (rs4810485 G/T) genetic models between the study and control groups. Linkage disequilibrium was found between the two SNPs and revealed four main haplotypes (i.e., C-G, C-T, T-G, and T-T), with the T-G haplotype having a significantly higher frequency in patients with ITP than in healthy controls, which conferred an increased risk of ITP development (OR, 2.349; 95% CI, 1.271–4.339;
P=0.006). They concluded that the
CD40 gene SNP rs1883832 represented an increased risk of developing ITP in the Egyptian population. The T-G haplotype is also considered a genetic risk model for ITP.
In a study by Yan
et al. [
12], which investigated the association between CD40 gene polymorphism and ITP in the Chinese population, the results revealed that the frequency of the CC genotype of rs1883832 in the ITP group was significantly higher than that in the control group (
P<0.05) and that the TT genotype and T allele in the ITP group were significantly lower than those in the control group (
P<0.05). Meanwhile, analysis of the rs4810485 locus showed no statistical difference in distribution between the two groups.
The discrepancy between the results could be a reflection of several factors, such as the limited number of cases, patient selection, genetic heterogeneity of various ethnic populations, and different age groups. Furthermore, it may be attributed to differences in disease pathogenesis and progression between the different age groups.
In this study, regarding the responsiveness to treatment among patients with ITP with regard to both SNPs, a statistically insignificant difference was detected, which could be attributed to the low number of nonresponsive cases (only one patient) (P>0.05).
In this study, the combined gene polymorphisms of rs1883832 C>T and rs4810485 G>T showed a highly statistically significant difference (P<0.001) between the patient and control groups, suggesting that carriers of both gene polymorphisms are at a higher risk of developing chronic ITP. Thus, the expression of CD40 plays an important role in the pathogenesis of chronic ITP in adolescent and adult patients and may contribute to the modification of the disease course. The rs4810485 and rs1883832 gene polymorphisms could be implemented as part of routine investigations for patients with ITP in the future to predict the disease course and hopefully help develop new promising therapeutic modalities for ITP. Further studies on the genes and pathways that modify the pathogenesis of ITP may help clarify the biology of ITP and suggest the possibility of novel testing for biomarkers of the disease or therapeutic targets.




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download