Abstract
Simultaneous myofibril and mitochondrial development is crucial for the cardiac differentiation of pluripotent stem cells (PSCs). Specifically, mitochondrial energy metabolism (MEM) development in cardiomyocytes is essential for the beating function. Although previous studies have reported that MEM is correlated with cardiac differentiation, the process and timing of MEM regulation for cardiac differentiation remain poorly understood. Here, we performed transcriptome analysis of cells at specific stages of cardiac differentiation from mouse embryonic stem cells (mESCs) and human induced PSCs (hiPSCs). We selected MEM genes strongly upregulated at cardiac lineage commitment and in a time-dependent manner during cardiac maturation and identified the protein-protein interaction networks. Notably, MEM proteins were found to interact closely with cardiac maturation-related proteins rather than with cardiac lineage commitment-related proteins. Furthermore, MEM proteins were found to primarily interact with cardiac muscle contractile proteins rather than with cardiac transcription factors. We identified several candidate MEM regulatory genes involved in cardiac lineage commitment (Cck, Bdnf, Fabp4, Cebpα, and Cdkn2a in mESC-derived cells, and CCK and NOS3 in hiPSC-derived cells) and cardiac maturation (Ppargc1α, Pgam2, Cox6a2, and Fabp3 in mESC-derived cells, and PGAM2 and SLC25A4 in hiPSC-derived cells). Therefore, our findings show the importance of MEM in cardiac maturation.
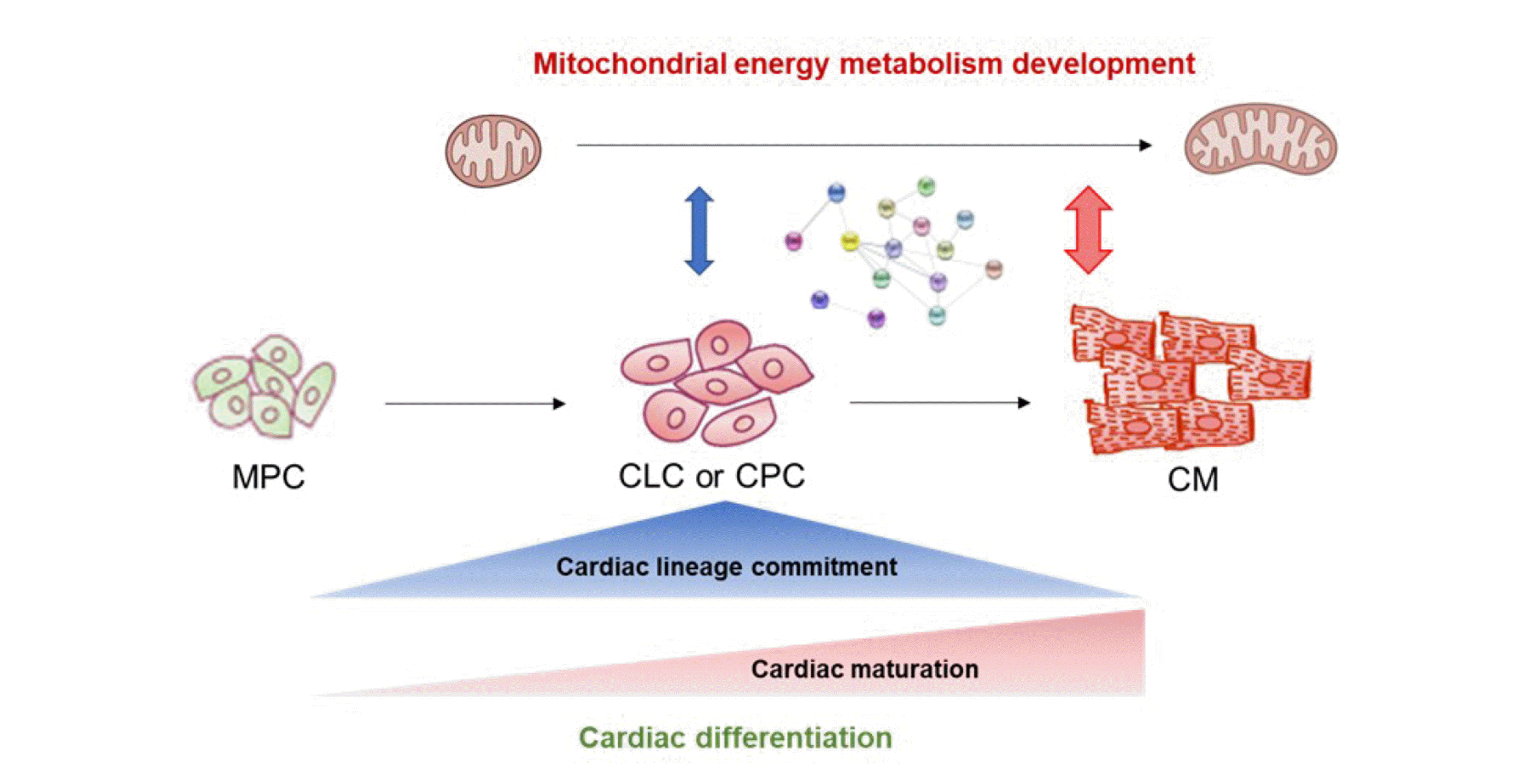
The concomitant development of myofibrils and mitochondria is a major process in cardiac differentiation from pluripotent stem cells (PSCs) [1]. In particular, the development of mitochondrial energy metabolism (MEM) in cardiomyocytes is crucial for the provision of energy necessary to support the beating function [2]. Therefore, the connection between MEM and transcriptional regulation is critical for cardiac differentiation, and the mitochondria-nucleus interaction is necessary for determining the signals that control cardiomyogenesis.
The process of cardiac differentiation of PSC-derived mesodermal cells generally involves cardiac lineage commitment and maturation. Cardiomyocyte proliferation and maturation occur simultaneously during the early stage of differentiation, including cardiac lineage commitment, whereas it decreases and the maturation process is dominant during the late stage of differentiation [3,4]. We previously reported a unique population of PSC-derived platelet-derived growth factor receptor-alpha (PDGFRα+) cardiac lineage-committed cells (CLCs), which are morphologically and functionally immature but actively proliferating compared to differentiated cardiomyocytes [5,6]. It has also been reported that cells expressing both kinase insert domain receptor (KDR) and PDGFRα belong to a population of cardiac progenitor cells (CPCs) with cardiomyogenic potential in human PSCs [7,8].
Although previous studies have reported that MEM is associated with cardiac differentiation, it is yet to be understood when and how MEM is regulated for this process. Here, we performed MEM transcriptome and protein-protein interaction (PPI) network analyses of cardiac lineage commitment and cardiac maturation using mouse embryonic stem cell (mESC)-derived PDGFRα+ CLCs and human induced PSC (hiPSC)-derived KDR+PDGFRα+ CPCs with those of mesodermal cells and differentiated cardiomyocytes.
The detailed methods used for the induction of vascular endothelial growth factor receptor 2 (Flk1+)-expressing mesodermal precursor cells (MPCs), PDGFRα+ CLCs, and alpha myosin heavy chain-expressing (αMHC+) cardiomyocytes from mESCs and KDR+PDGFRα+ cells and cardiomyocytes from hiPSCs have been described previously [5,8].
We obtained raw microarray data from the Gene Expression Omnibus database (accession numbers GSE65791 and GSE90000) for comparative analysis, which were selected based on the Gene Ontology (GO) database and Qiagen PCR-array gene set. GO annotations were searched using QuickGO (https://www.ebi.ac.uk/QuickGO/). The MEM gene category included the following GO annotations and PCR arrays: GO-0006119 (oxidative phosphorylation), GO-0005739 (mitochondria), GO-0007005 (mitochondrial organization), GO-0031966 (mitochondrial membrane), PAMM-006ZA (glucose metabolism), PAMM-007ZA (fatty acid metabolism), and PAMM-008ZA (MEM). Differentially expressed genes were analyzed using the Microsoft Excel-based differentially expressed gene analysis (ExDEGA) software (EBIOGEN, Seoul, Korea).
PPI analysis was performed using Cytoscape v3.7.0 and STRING software (https://string-db.org/). Significantly upregulated genes were defined as those with a fold change ≥ 2, normalized data (log 2) of 4, and p < 0.05. For mESC-derived cells, MEM genes that were specifically upregulated in PDGFRα+ CLCs at cardiac lineage commitment compared to that in Flk1+ MPCs and αMHC+ cardiomyocytes and were continuously upregulated in a time-dependent manner during cardiac maturation were selected for PPI analysis. In hiPSC-derived cells, the MEM genes selected for PPI analysis were those that were specifically upregulated in the KDR+PDGFRα+ CPCs at cardiac lineage commitment compared to that in KDR+PDGFRα+ cells on day 4 and in cardiomyocytes on day 19, and were continuously upregulated during cardiac maturation. We applied the selected MEM genes to PPI networks at cardiac lineage commitment and during cardiac maturation from our previous study (Supplementary Figs. 1 and 2) and to newly constructed MEM PPI networks [3].
Human embryonic stem cell (hESC)-derived cardiac differentiation was induced as previously described [9]. For cardiac lineage induction, H9 hESCs (WiCell, Madison, WI, USA) were plated onto Matrigel (Corning, New York, NY, USA)-coated 6 well plate at a density of 0.5 × 106 cells/well and cultured in mTeSR1 (STEMCELL Technologies, Vancouver, BC, Canada) supplemented with cGMP for 2–3 days. Three days before cardiomyocyte differentiation, hESCs were plated on Matrigel-coated 6 well plate at a density of 0.6 × 106 cells/well and cultured in mTeSR1 supplemented with 5 μM Y27632 (Sigma-Aldrich, St. Louis, MO, USA). The culture medium was replaced with RPMI+B27 medium (RPMI1640 with B27 supplement without insulin; Gibco, Grand Island, NY, USA) supplemented with CHIR99021 (10 μM, S1263; Selleckchem, Houston, TX, USA) for 3 days. The culture medium was subsequently replaced with RPMI+B27 supplemented with IWP2 (5 μM; Tocris, Bristol, UK), followed by culture for 5 days. On day 8, the culture medium was replaced with RPMI+B27. The medium was changed every 1–2 days. Beating cardiomyocytes were observed on days 9–10. Cardiomyocytes were chemically sorted by lactate (Wako, Richmond, VA, USA).
Total RNA was extracted using the TRIzol RNA extraction kit (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s instructions. Total RNA was reverse transcribed into cDNA using the SuperScript VILO cDNA Synthesis Kit (Invitrogen). cDNA was used for qPCR using TOPreal qPCR 2X Pre-MIX (Enzynomics, Daejeon, Korea) and 7500 Real-Time PCR Systems (Applied Biosystems, Foster City, CA, USA) with the indicated primers (Supplementary Table 1). Primers were made by using Primer3 (https://bioinfo.ut.ee/primer3-0.4.0/). GAPDH was used as a reference gene, and the expression levels are presented as values relative to those in the control using the ΔΔCt method. Significant differences between means were determined by one-way analysis of variance, followed by the Student–Newman–Keuls test. Statistical significance was set at p < 0.05.
During cardiac differentiation from PSCs, we defined the specific stages as mesodermal induction, cardiac lineage commitment and maturation. At the stage of cardiac lineage commitment, we sorted out mESC-derived PDGFRα+ CLCs and hiPSC-derived KDR+PDGFRα+ CPCs, which are immature without beating property but actively proliferating compared to matured cardiomyocytes. At the stage of cardiac maturation, we sorted out mESC-derived αMHC+ cardiomyocytes and hiPSC-derived cardiomyocytes, which have beating property. The specific time points of cell sorting for microarray analysis during cardiac differentiation are shown in Fig. 1A and 1B.
Among mESC-derived cells, 16 MEM genes were significantly upregulated in PDGFRα+ CLCs at cardiac lineage commitment compared to that in Flk1+ MPCs and αMHC+ cardiomyocytes (Fig. 1C). There were 23 continuously upregulated MEM genes during cardiac maturation (Fig. 1D).
In the hiPSC-derived cells, 23 MEM genes in the KDR+PDGFRα+ CPCs were significantly upregulated at cardiac lineage commitment compared to that in KDR+PDGFRα+ cells on day 4 and in cardiomyocytes on day 19 (Fig. 1E). There were 28 MEM genes that underwent continuous time-dependent upregulation during cardiac maturation (Fig. 1F).
We conducted a PPI analysis to gain insights into the role of MEM in cardiac lineage commitment and maturation. Additionally, we identified novel candidate MEM regulatory genes for cardiac cell differentiation from the PPI networks.
In mESC-derived cells, the MEM and cardiac lineage commitment PPI networks had 50 PPIs, whereas the MEM and cardiac maturation PPI networks had 97 PPIs (Fig. 2A). The genes constituting the MEM PPI networks of the mESC-derived cells are listed in Supplementary Table 2. During cardiac lineage commitment, cholecystokinin (Cck) and brain-derived neurotrophic factor (BDNF) are connected with Wnt family member 2, fatty acid-binding protein 4 (Fabp4), and CCAAT/enhancer binding protein alpha (Cebpα), which are connected to myocyte-specific enhancer factor 2C (Fig. 2B). During cardiac maturation, peroxisome proliferator-activated receptor gamma coactivator 1 alpha (Ppargc1α), phosphoglycerate mutase 2 (Pgam2), cytochrome c oxidase subunit 6A2 (Cox6a2), and fatty acid-binding protein 3 (Fabp3) are directly connected to cardiac muscle contractile proteins (Fig. 2C).
In hiPSC-derived cells, the MEM metabolism and cardiac lineage commitment PPI networks had ten PPIs, whereas the MEM and cardiac maturation PPI networks had 43 PPIs (Fig. 3A). Genes constituting the MEM PPI networks of hiPSC-derived cells are listed in Supplementary Table 3. During cardiac lineage commitment, CCK and nitric oxide synthase 3 (NOS3) are connected to adrenoceptor alpha 1A (Fig. 3B). During cardiac maturation, PGAM2 and solute carrier family 25 member 4 (SLC25A4) are directly connected to cardiac muscle contractile proteins (Fig. 3C).
Collectively, the MEM PPI network showed greater development during cardiac maturation than during cardiac lineage commitment. Furthermore, MEM proteins primarily interact with cardiac muscle contractile proteins rather than with cardiac transcription factors such as TBX5, HAND2, MEF2C, and NKX2-5. These data indicate that MEM is important for cardiac maturation rather than for cardiac lineage commitment during the cardiac differentiation of PSCs.
To confirm the expression of CCK, NOS3, PGAM2, and SLC25A4, which were directly connected with cardiac lineage commitment and maturation related genes from hiPSC-derived PPI networks during cardiac differentiation, we induced cardiac differentiation using hESCs. Undifferentiated hESCs were observed on day –3 to 0, mesodermal cells on day 3, cardiac progenitor cells on day 8, and cardiomyocytes on day 16 upon the differentiation of hESCs (Fig. 4A). Quantitative polymerase chain reaction (qPCR) was performed at each developmental stage. Consistent with the findings of the transcriptome analysis, with the exception of CCK, the other genes showed changes in expression levels; the mRNA expression level of NOS3 was specifically upregulated in the CPCs, and those of PGAM2 and SLC25A4 were specifically upregulated in the cardiomyocytes (Fig. 4B, C). We interpreted that the several inconsistencies of gene expression at each developmental stage between microarray and qPCR data might be caused by different cardiac differentiation protocols.
In the present study, we explored the landscape of MEM PPI networks during cardiac lineage commitment and maturation. PPI analysis revealed that MEM proteins interact more closely with cardiac maturation-related proteins than with cardiac lineage commitment-related proteins (Fig. 5). In addition, MEM proteins primarily interact with cardiac muscle contractile proteins rather than with cardiac transcription factors. Therefore, MEM is important for cardiac maturation rather than cardiac lineage commitment. We also identified several candidate MEM regulatory genes involved in cardiac lineage commitment (Cck, Bdnf, Fabp4, Cebpα, and Cdkn2a in mESC-derived cells, and CCK and NOS3 in hiPSC-derived cells) and cardiac maturation (Ppargc1α, Pgam2, Cox6a2, and Fabp3 in mESC-derived cells, and PGAM2 and SLC25A4 in hiPSC-derived cells).
Conventionally, cellular metabolism is considered a passenger for PSCs and differentiated cells for proliferation, self-renewal, and differentiation [2,10]. However, this notion is changing because cellular metabolism drives differentiation, including fate determination and maturation [1,2]. During cardiac differentiation from PSCs, the mitochondria develop, and the key process in cellular metabolism changes from glycolysis and glutamine oxidation to lactate and fatty acid oxidation [1]. Based on these developmental metabolic profiles, a large-scale metabolic sorting system was developed for hPSC-derived cardiomyocytes [11]. Furthermore, several studies have demonstrated that mitochondrial metabolic regulation affects cardiac differentiation from PSCs [1,12]. We previously reported that the activation of mitochondrial oxidative metabolism via mitochondrial permeability transition pore inhibition by cyclosporin A enhances cardiac differentiation from PSCs [13]. However, the specific time point and detailed mechanism underlying the regulation of cardiac differentiation by MEM remain unknown.
In addition, we have previously shown the transcriptome profiles and the PPI networks at cardiac lineage commitment comparing with those of mesodermal cells and differentiated cardiomyocytes [3]. Similarly, in the present study, we performed MEM PPI analysis at cardiac lineage commitment and during cardiac maturation. Consistent with previous findings, our findings indicate that MEM possibly plays a greater role in cardiac maturation than in fate determination [2]. Molecular mechanisms triggered by metabolic stimuli may contribute to cardiomyocyte cell cycle exit and maturation. Indeed, previous studies have shown that metabolic stimuli, including molecules, such as fatty acids and hormones, and processes, such as glucose removal, can improve PSC-derived cardiomyocyte maturation [14-16]. The mitochondria undergo maturation during the perinatal period, transitioning from small, fragmented organelles to large organelles with developed cristae capable of producing sufficient ATP to support the beating function of the postnatal heart [2]. Mitochondrial maturation may not merely be a response to cardiac maturation; rather, the mitochondria may facilitate molecular processes by responding to nutrient signals, dynamically altering the respiratory capacity and ROS production, and initiating transcriptional and epigenetic changes triggering cardiac maturation [2].
In the present study, we selected several candidate MEM regulatory genes involved in cardiac lineage commitment and during cardiac maturation. These genes have been associated with cardiac development and differentiation in previous studies, which have reported that CCK is expressed in the embryonic murine heart, whereas pro-CCK is expressed in the adult mammalian heart [17,18]. However, the exact function of CCK is yet to be confirmed. Furthermore, the C/EBP family of transcription factors is activated in the epicardium in response to both developmental and injury signals to establish a transcriptional code for embryonic gene expression [19]. Another study showed that Betrofin3, an agonist of the BDNF peptide, exerted a robust pro-cardiomyogenic effect on embryonic stem cells via the TrkB receptor [20]. Interestingly, NOS3 was also expressed in the embryonic heart at E9.5, with the expression level remaining high till E13.5 and decreasing from E14.5, in both the atria and ventricles [21]. NOS3 deficiency results in congenital septal defects, cardiac hypertrophy, and postnatal heart failure [21]. Additionally, NOS3 is important in the development of the major coronary artery, myocardial capillaries, and aortic valve [21]. Previous studies have reported that FABP3 is expressed at high levels in patients with ventricular septal defects, and FABP3 knockdown impairs cardiac development in zebrafish through the retinoic acid signaling pathway [22]. In addition, PGC1/PPAR drives PSC-derived cardiomyocyte maturation at the single-cell level via YAP1, SF3B2, and the PPARGC1α activator ZLN005 also promotes hESC-derived cardiomyocyte maturation [23,24]. Another study reported that the knockout of Slc25a4 mice with mitochondrial energy deficiency led to the development of a distinctive concentric dilated cardiomyopathy with cardiac dysfunction mediated via increased cytoplasmic cytochrome c levels and caspase 3 activation [25]. However, these studies did not investigate the detailed molecular and cellular mechanism of action of genes involved in MEM. Further studies using iPSC-derived cardiomyocytes with genetic knockdown are warranted to clarify the exact molecular and cellular mechanisms underlying the action of MEM regulatory genes involved in cardiac lineage commitment and maturation. Furthermore, the molecular and cellular mechanisms of MEM regulatory genes involved in cardiac lineage commitment and maturation may help generate abundant mature cardiomyocytes for cardiac regeneration.
In conclusion, we demonstrated that MEM is regulated differently in each stage of cardiac differentiation; in particular, PPI increases rapidly during cardiac maturation rather than during cardiac lineage commitment. These results support the theory that MEM may exert a greater impact on the cardiac maturation stage in creating cardiac functional integrity for beating. Our findings provide valuable insights into how MEM can affect cardiac lineage commitment and maturation. The findings may also help develop alternative strategies for improving cardiac differentiation and maturation for generating mature cardiomyocytes in the field of regenerative medicine.
Supplementary data including two figures and three tables can be found with this article online at https://doi.org/10.4196/kjpp.2022.26.5.357.
Notes
FUNDING
This research was funded by the Basic Science Research Program through the National Research Foundation (NRF) of Korea funded by the Ministry of Education (2017R1D1A3B03034465), the NRF of Korea funded by the Ministry of Science and ICT (MSIT) (2020R1C1C1015104), and the Basic Research Lab Program through the NRF of Korea funded by the MSIT (2020R1A4A1018943).
REFERENCES
1. Morita Y, Tohyama S. 2020; Metabolic regulation of cardiac differentiation and maturation in pluripotent stem cells: a lesson from heart development. JMA J. 3:193–200. DOI: 10.31662/jmaj.2020-0036. PMID: 33150253. PMCID: PMC7590396.

2. Garbern JC, Lee RT. 2021; Mitochondria and metabolic transitions in cardiomyocytes: lessons from development for stem cell-derived cardiomyocytes. Stem Cell Res Ther. 12:177. DOI: 10.1186/s13287-021-02252-6. PMID: 33712058. PMCID: PMC7953594. PMID: 26365eb57d8140289dd56323c91b3762.

3. Cho SW, Kim HK, Sung JH, Han J. 2021; Stage specific transcriptome profiles at cardiac lineage commitment during cardiomyocyte differentiation from mouse and human pluripotent stem cells. BMB Rep. 54:464–469. DOI: 10.5483/BMBRep.2021.54.9.046. PMID: 34120677. PMCID: PMC8505231.

4. Kamps JA, Krenning G. 2016; Micromanaging cardiac regeneration: targeted delivery of microRNAs for cardiac repair and regeneration. World J Cardiol. 8:163–179. DOI: 10.4330/wjc.v8.i2.163. PMID: 26981212. PMCID: PMC4766267.

5. Hong SP, Song S, Cho SW, Lee S, Koh BI, Bae H, Kim KH, Park JS, Do HS, Im I, Heo HJ, Ko TH, Park JH, Youm JB, Kim SJ, Kim I, Han J, Han YM, Koh GY. 2017; Generation of PDGFRα+ cardioblasts from pluripotent stem cells. Sci Rep. 7:41840. DOI: 10.1038/srep41840. PMID: 28165490. PMCID: PMC5292955.

6. Hong SP, Song S, Lee S, Jo H, Kim HK, Han J, Park JH, Cho SW. 2019; Regenerative potential of mouse embryonic stem cell-derived PDGFRα+ cardiac lineage committed cells in infarcted myocardium. World J Stem Cells. 11:44–54. DOI: 10.4252/wjsc.v11.i1.44. PMID: 30705714. PMCID: PMC6354102.

7. Kattman SJ, Witty AD, Gagliardi M, Dubois NC, Niapour M, Hotta A, Ellis J, Keller G. 2011; Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 8:228–240. DOI: 10.1016/j.stem.2010.12.008. PMID: 21295278.

8. Takeda M, Kanki Y, Masumoto H, Funakoshi S, Hatani T, Fukushima H, Izumi-Taguchi A, Matsui Y, Shimamura T, Yoshida Y, Yamashita JK. 2018; Identification of cardiomyocyte-fated progenitors from human-induced pluripotent stem cells marked with CD82. Cell Rep. 22:546–556. DOI: 10.1016/j.celrep.2017.12.057. PMID: 29320747.

9. Lian X, Zhang J, Azarin SM, Zhu K, Hazeltine LB, Bao X, Hsiao C, Kamp TJ, Palecek SP. 2013; Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions. Nat Protoc. 8:162–175. DOI: 10.1038/nprot.2012.150. PMID: 23257984. PMCID: PMC3612968.

10. Porter GA Jr, Hom J, Hoffman D, Quintanilla R, de Mesy Bentley K, Sheu SS. 2011; Bioenergetics, mitochondria, and cardiac myocyte differentiation. Prog Pediatr Cardiol. 31:75–81. DOI: 10.1016/j.ppedcard.2011.02.002. PMID: 21603067. PMCID: PMC3096664.

11. Tohyama S, Hattori F, Sano M, Hishiki T, Nagahata Y, Matsuura T, Hashimoto H, Suzuki T, Yamashita H, Satoh Y, Egashira T, Seki T, Muraoka N, Yamakawa H, Ohgino Y, Tanaka T, Yoichi M, Yuasa S, Murata M, Suematsu M, et al. 2013; Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes. Cell Stem Cell. 12:127–137. DOI: 10.1016/j.stem.2012.09.013. PMID: 23168164.

12. Heo HJ, Kim HK, Youm JB, Cho SW, Song IS, Lee SY, Ko TH, Kim N, Ko KS, Rhee BD, Han J. 2016; Mitochondrial pyruvate dehydrogenase phosphatase 1 regulates the early differentiation of cardiomyocytes from mouse embryonic stem cells. Exp Mol Med. 48:e254. DOI: 10.1038/emm.2016.70. PMID: 27538372. PMCID: PMC5007642.

13. Cho SW, Park JS, Heo HJ, Park SW, Song S, Kim I, Han YM, Yamashita JK, Youm JB, Han J, Koh GY. 2014; Dual modulation of the mitochondrial permeability transition pore and redox signaling synergistically promotes cardiomyocyte differentiation from pluripotent stem cells. J Am Heart Assoc. 3:e000693. DOI: 10.1161/JAHA.113.000693. PMID: 24627421. PMCID: PMC4187507.

14. Yang X, Rodriguez ML, Leonard A, Sun L, Fischer KA, Wang Y, Ritterhoff J, Zhao L, Kolwicz SC Jr, Pabon L, Reinecke H, Sniadecki NJ, Tian R, Ruohola-Baker H, Xu H, Murry CE. 2019; Fatty acids enhance the maturation of cardiomyocytes derived from human pluripotent stem cells. Stem Cell Reports. 13:657–668. DOI: 10.1016/j.stemcr.2019.08.013. PMID: 31564645. PMCID: PMC6829750.

15. Feyen DAM, McKeithan WL, Bruyneel AAN, Spiering S, Hörmann L, Ulmer B, Zhang H, Briganti F, Schweizer M, Hegyi B, Liao Z, Pölönen RP, Ginsburg KS, Lam CK, Serrano R, Wahlquist C, Kreymerman A, Vu M, Amatya PL, Behrens CS, et al. 2020; Metabolic maturation media improve physiological function of human iPSC-derived cardiomyocytes. Cell Rep. 32:107925. DOI: 10.1016/j.celrep.2020.107925. PMID: 32697997. PMCID: PMC7437654.

16. Mills RJ, Titmarsh DM, Koenig X, Parker BL, Ryall JG, Quaife-Ryan GA, Voges HK, Hodson MP, Ferguson C, Drowley L, Plowright AT, Needham EJ, Wang QD, Gregorevic P, Xin M, Thomas WG, Parton RG, Nielsen LK, Launikonis BS, James DE, et al. 2017; Functional screening in human cardiac organoids reveals a metabolic mechanism for cardiomyocyte cell cycle arrest. Proc Natl Acad Sci U S A. 114:E8372–E8381. DOI: 10.1073/pnas.1707316114. PMID: 28916735. PMCID: PMC5635889.

17. Lay JM, Gillespie PJ, Samuelson LC. 1999; Murine prenatal expression of cholecystokinin in neural crest, enteric neurons, and enteroendocrine cells. Dev Dyn. 216:190–200. DOI: 10.1002/(SICI)1097-0177(199910)216:2<190::AID-DVDY9>3.0.CO;2-K. PMID: 10536058.

18. Goetze JP, Johnsen AH, Kistorp C, Gustafsson F, Johnbeck CB, Rehfeld JF. 2015; Cardiomyocyte expression and cell-specific processing of procholecystokinin. J Biol Chem. 290:6837–6843. DOI: 10.1074/jbc.M114.622670. PMID: 25627687. PMCID: PMC4358109.

19. Huang GN, Thatcher JE, McAnally J, Kong Y, Qi X, Tan W, DiMaio JM, Amatruda JF, Gerard RD, Hill JA, Bassel-Duby R, Olson EN. 2012; C/EBP transcription factors mediate epicardial activation during heart development and injury. Science. 338:1599–1603. DOI: 10.1126/science.1229765. PMID: 23160954. PMCID: PMC3613149.

20. Xu R, inivasan SP Sr, Sureshkumar P, Nembo EN, Schäfer C, Semmler J, Matzkies M, Albrechtsen M, Hescheler J, Nguemo F. 2015; Effects of synthetic neural adhesion molecule mimetic peptides and related proteins on the cardiomyogenic differentiation of mouse embryonic stem cells. Cell Physiol Biochem. 35:2437–2450. DOI: 10.1159/000374044. PMID: 25967873.

21. Liu Y, Feng Q. 2012; NOing the heart: role of nitric oxide synthase-3 in heart development. Differentiation. 84:54–61. DOI: 10.1016/j.diff.2012.04.004. PMID: 22579300.

22. Wang X, Zhou L, Jin J, Yang Y, Song G, Shen Y, Liu H, Liu M, Shi C, Qian L. 2013; Knockdown of FABP3 impairs cardiac development in Zebrafish through the retinoic acid signaling pathway. Int J Mol Sci. 14:13826–13841. DOI: 10.3390/ijms140713826. PMID: 23823803. PMCID: PMC3742220.

23. Murphy SA, Miyamoto M, Kervadec A, Kannan S, Tampakakis E, Kambhampati S, Lin BL, Paek S, Andersen P, Lee DI, Zhu R, An SS, Kass DA, Uosaki H, Colas AR, Kwon C. 2021; PGC1/PPAR drive cardiomyocyte maturation at single cell level via YAP1 and SF3B2. Nat Commun. 12:1648. DOI: 10.1038/s41467-021-21957-z. PMID: 33712605. PMCID: PMC7955035. PMID: e529861439104233888f042c1cc8bff3.

24. Liu Y, Bai H, Guo F, Thai PN, Luo X, Zhang P, Yang C, Feng X, Zhu D, Guo J, Liang P, Xu Z, Yang H, Lu X. 2020; PGC-1α activator ZLN005 promotes maturation of cardiomyocytes derived from human embryonic stem cells. Aging (Albany NY). 12:7411–7430. DOI: 10.18632/aging.103088. PMID: 32343674. PMCID: PMC7202542.

25. Narula N, Zaragoza MV, Sengupta PP, Li P, Haider N, Verjans J, Waymire K, Vannan M, Wallace DC. 2011; Adenine nucleotide translocase 1 deficiency results in dilated cardiomyopathy with defects in myocardial mechanics, histopathological alterations, and activation of apoptosis. JACC Cardiovasc Imaging. 4:1–10. DOI: 10.1016/j.jcmg.2010.06.018. PMID: 21232697. PMCID: PMC4023693.

Fig. 1
Significantly upregulated MEM-related genes at cardiac lineage commitment and during cardiac maturation.
(A) Sampling time points of mESC-derived cells: Flk1+ MPCs on day 4.5, PDGFRα+ CLCs on day 6, and αMHC+ cardiomyocytes on day 10.5. (B) Sampling time points of hiPSC-derived cells: KDR+PDGFRα+ cells on days 4 and 5 and cardiomyocytes on day 19. (C) Significantly upregulated MEM genes in mESC-derived PDGFRα+ CLCs at cardiac lineage commitment. (D) MEM genes with continuous time-dependent upregulation during cardiac maturation in mESC-derived cells. (E) Significantly upregulated MEM genes in hiPSC-derived KDR+PDGFRα+ CPCs at cardiac lineage commitment. (F) MEM genes undergoing continuous time-dependent upregulation during cardiac maturation in hiPSC-derived cells. αMHC, alpha-myosin heavy chain; CLC, cardiac lineage-committed cell; CPC, cardiac progenitor cell; CM, cardiomyocyte; Flk1, vascular endothelial growth factor receptor 2; hiPSC, human induced pluripotent stem cell; KDR, kinase insert domain receptor; MEM, mitochondrial energy metabolism; mESC, mouse embryonic stem cell; MPC, mesodermal precursor cell; PDGFRα, platelet-derived growth factor receptor alpha.
Fig. 2
Protein-protein interaction networks from mESC-derived cells.
(A) PPI networks of MEM and cardiac lineage commitment and maturation in mESC-derived cells. (B) PPI network connection of MEM regulatory genes and cardiac lineage commitment-related genes in mESC-derived cells. (C) PPI network connection of MEM regulatory genes and cardiac maturation-related genes in mESC-derived cells. The red and blue colors mean the gene expression changes at the stages of cardiac maturation and lineage commitment. The circle means cardiac lineage commitment related gene, the triangle means cardiac maturation related gene, and the diamond is mitochondria related gene. The gray line is the interactions among the genes except interactions of mitochondria-cardiac lineage commitment related genes (green line) or mitochondria-cardiac maturation related genes (red line). mESC, mouse embryonic stem cell; PPI, protein-protein interaction; MEM, mitochondrial energy metabolism.
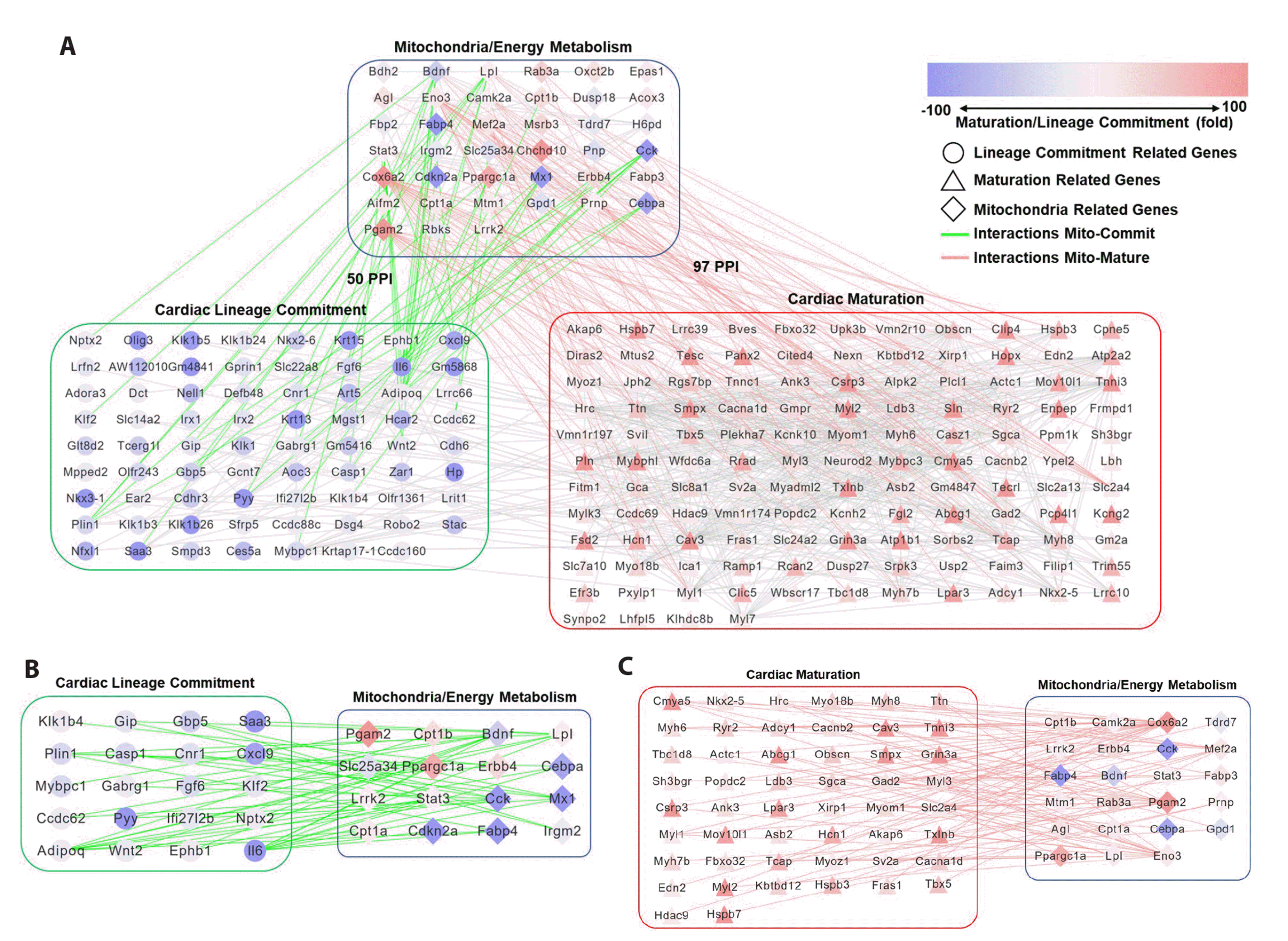
Fig. 3
Protein-protein interaction networks from hiPSC-derived cells.
(A) PPI networks of MEM and cardiac lineage commitment and maturation in hiPSC-derived cells. (B) PPI network connection of MEM regulatory genes and cardiac lineage commitment-related genes in hiPSC-derived cells. (C) PPI network connection of MEM regulatory genes and cardiac maturation-related genes in hiPSC-derived cells. The red and blue colors mean the gene expression changes at the stages of cardiac maturation and lineage commitment. The circle means cardiac lineage commitment related gene, the triangle means cardiac maturation related gene, and the diamond is mitochondria related gene. The gray line is the interactions among the genes except interactions of mitochondria-cardiac lineage commitment related genes (green line) or mitochondria-cardiac maturation related genes (red line). hiPSC, human induced pluripotent stem cell; PPI, protein-protein interaction; MEM, mitochondrial energy metabolism.
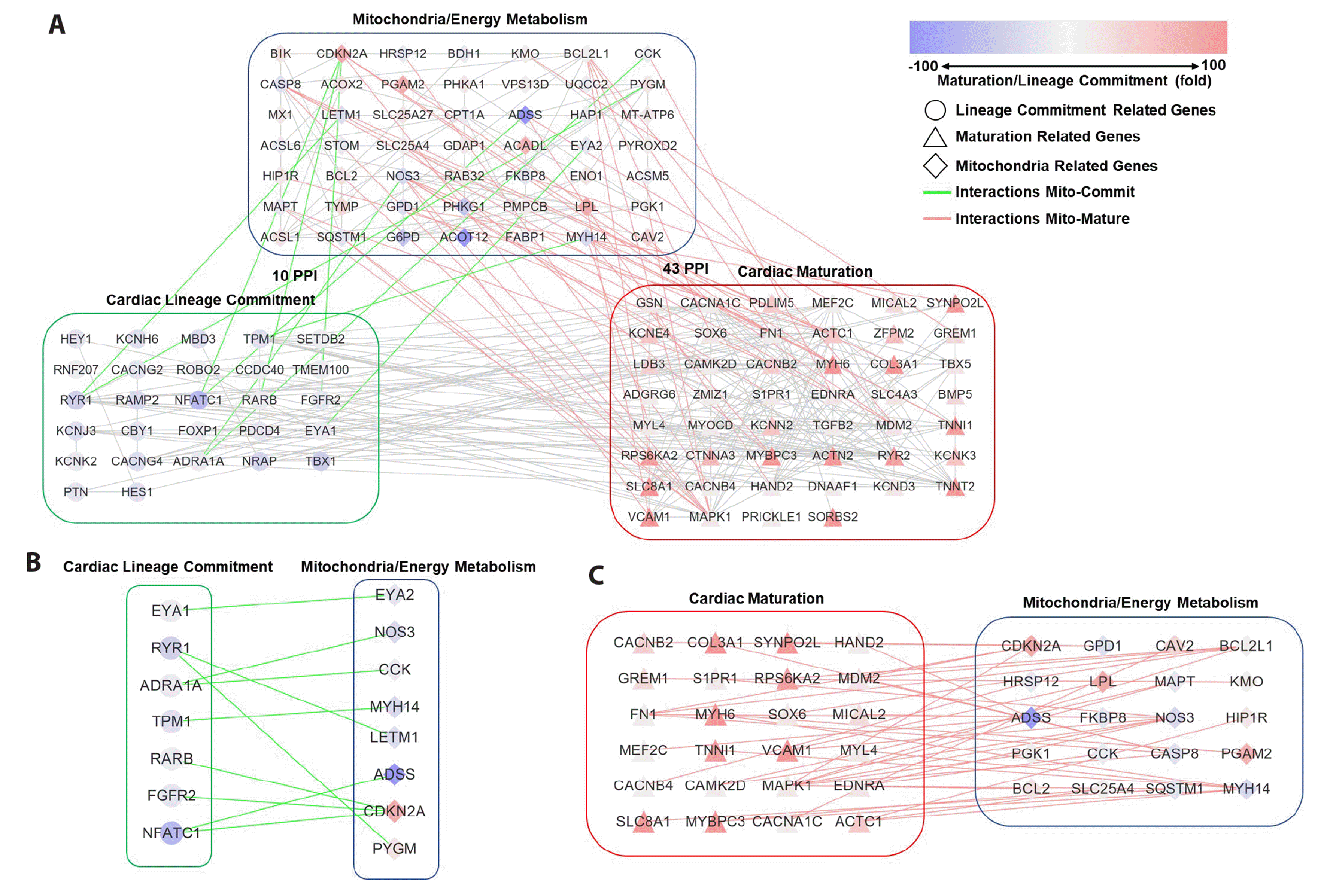
Fig. 4
Confirmation of MEM regulatory gene candidates during cardiac differentiation from hESCs.
(A) Schematic cardiomyocyte differentiation protocol and sampling time points of the hESC-derived cells: mesodermal cells on day 3, cardiac progenitor cells on day 8, and cardiomyocytes on day 16, formed from the differentiation of H9 hESCs (scale bars, 50 μm). (B) The expression level of CCK, NOS3, PGAM2, and SLC25A4 at each developmental stage from microarray. (C) The expression level of CCK, NOS3, PGAM2, and SLC25A4 at each developmental stage from qPCR. *p-value < 0.005. RBIn, RPMI1640+B27 without Insulin; RB+, RPMI1640+B27 supplement. MEM, mitochondrial energy metabolism; hESC, human embryonic stem cell; CCK, cholecystokinin; NOS3, nitric oxide synthase 3; SLC25A4, solute carrier family 25 member 4; qPCR, quantitative polymerase chain reaction.




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download