CD36 is expressed in a variety of cells, including platelets, monocytes, macrophages, and endothelial and epithelial cells [
1,
2]. There are two types of CD36 deficiencies: type I, which lacks CD36 on both platelets and monocytes, and type II, which lacks CD36 on platelets alone. Only type I individuals are at risk of developing isoantibodies against CD36 [
3]. The prevalence of type I varies among different ethnic groups; it is ~3.0% in the African population, 1.0% in the Japanese population, and ~0.5% in the Chinese population [
4-
6]. Anti-CD36 isoantibodies (known as anti-Nak
a) can cause different immune-mediated bleeding disorders, including platelet transfusion refractoriness (PTR), fetal neonatal alloimmune thrombocytopenia (FNAIT), and posttransfusion purpura (PTP) [
7-
9]. Transfusion-related acute lung injury (TRALI) associated with anti-CD36 antibodies (Abs) has also been reported [
10].
Various methods have been developed for anti-CD36 Ab detection, including flow cytometry and antigen capture assays using platelets, such as the monoclonal antibody (mAb) immobilization of platelet antigens (MAIPA) assay, or stably transfected cell lines as the target [
4,
6,
11-
13]. Additionally, a solid-phase ELISA using purified CD36 is commercially available (PakPlus; Immucor GTI Diagnostics, Inc., Waukesha, WI, USA). In contrast to protein-based methods, antigen capture assays allow the testing of recipient serum with donor platelets carrying native, individual CD36 antigens and
vice versa (crossmatch). However, these assays depend on the use of a capture mAb against CD36 [
14]. Due to competitive inhibition of CD36 mAb by human anti-CD36 Abs in patients, the rate of false-negative reactions is high [
11,
13]. To address this, we developed a panel of mAbs against CD36 and selected noncompetitive mAbs for anti-CD36 Ab detection by an antigen capture assay.
Male CD36–/– mice (B6.129S1-Cd36tm1Mfe/J; Jackson Laboratory, Bar Harbor, ME, USA) were immunized with 5×106 HEK293T cells expressing mouse CD36 intraperitoneally three times on days 1, 8, 15; a booster was administered once on day 19. Splenocytes from the immunized mice were fused with SP2/0-Ag14 mouse myeloma cells (American Type Culture Collection, Manassas, VA, USA). Hybridomas were selected on hypoxanthine-aminopterin-thymidine medium (Life Technologies, Darmstadt, Germany) and screened by flow cytometry (BD FACSCanto II; BD Biosciences, San Jose, CA, USA) using HEK293T cells expressing mouse or human CD36. The study was conducted from July 2014 to May 2020 in the Animal Center of Sun Yat-Sen University, Guangzhou, China, following approval by the University’s Animal Ethical and Welfare Committee (IACUC-2014-0303).
Seven out of 25 clones that produced strong anti-CD36 Abs (termed GZ-3, GZ-13, GZ-70, GZ-143, GZ-413, GZ-507, and GZ-608) were selected. Flow cytometry results showed that all seven mAbs reacted with both mouse and human CD36 expressed on HEK293T cells, underscoring the high sequence similarity (85.0%) between mouse and human CD36 [
15]. In the control experiment, no reaction was observed with HEK293T cells transfected with a CD36 vector that carries the C220T variant, which leads to a premature stop codon [
6]. These results were confirmed by flow cytometry using normal platelets and platelets from CD36-negative individuals.
Fig. 1 shows the flow cytometry results for GZ-70 and GZ-608 mAbs.
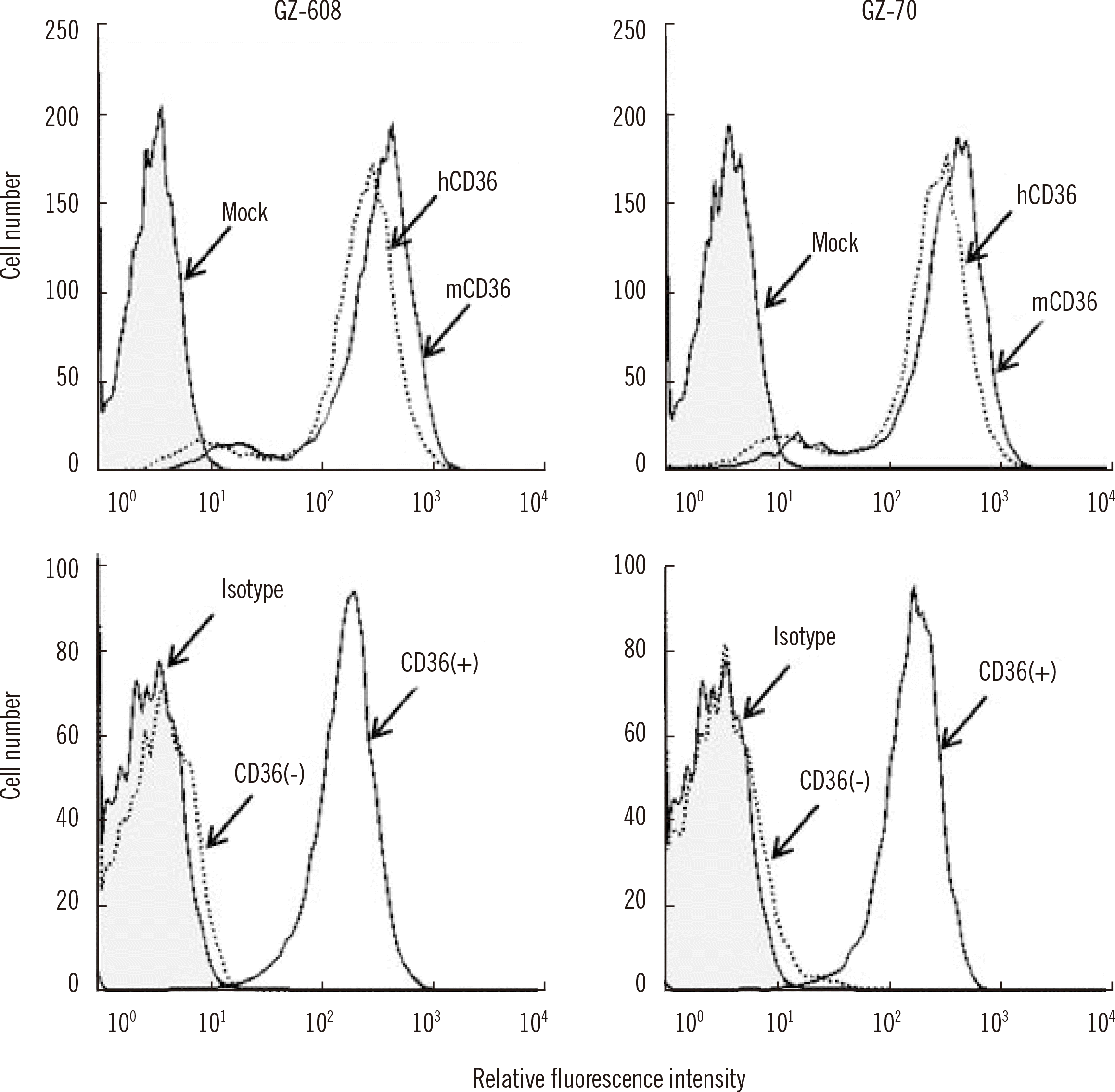 | Fig. 1Flow cytometry analysis of monoclonal antibodies (mAbs) against CD36 using transfected HEK293T cells and platelets. Upper panels: mAbs (GZ-608 and GZ-70) were incubated with mock, hCD36, and mCD36. Bound antibodies were detected using fluorescence-conjugated goat anti-mouse IgG and analyzed using flow cytometry. Lower panels: CD36+ platelets from a healthy blood donor and CD36− platelets from a CD36-deficient donor were incubated with GZ-608 or GZ-70 mAb. “Mock” stands for C220T variant CD36 construct-transfected HEK293T cells, “hCD36” for human CD36-transfected HEK293T cells, “mCD36” for mouse CD36-transfected HEK293T cells, and “Isotype” for mouse IgG1. 
|
Fourteen anti-CD36 sera from eight patients with PTR, four with FNAIT, and two healthy donors were first tested using the PakPlus ELISA kit. Informed consent was obtained from all individuals. These sera were analyzed by MAIPA assays using the reference anti-CD36 mAb FA6-152 and our mAbs against CD36. Briefly, 4×10
7 platelets were incubated with 25 µL of serum at 37°C for 30 minutes. After washing, 10 µL of anti-CD36 mAb (20 µg/mL) was added, and the samples were incubated at 37°C for 30 minutes. Then, the platelets were washed and solubilized, and bound anti-CD36 Abs were detected using 100 µL of o-phenylenediamine (Dako, Glostrup, Denmark) as a substrate [
16]. After 15 minutes, the reaction was stopped with 2.5 N H
2SO
4, and the optical density at 492 nm was measured using an ELISA reader (Multiskan FC; Thermo Scientific, Shanghai, China).
Fig. 2A shows the reactivity of one anti-CD36 serum (serum number 8;
Table 1) with different anti-CD36 mAbs. Only 3/7 mAbs (GZ-13, GZ-70, and GZ-608) were capable of detecting the anti-CD36 Abs present in this serum (
Fig. 2A). False-negative results were obtained for five mAbs, including FA6-152.
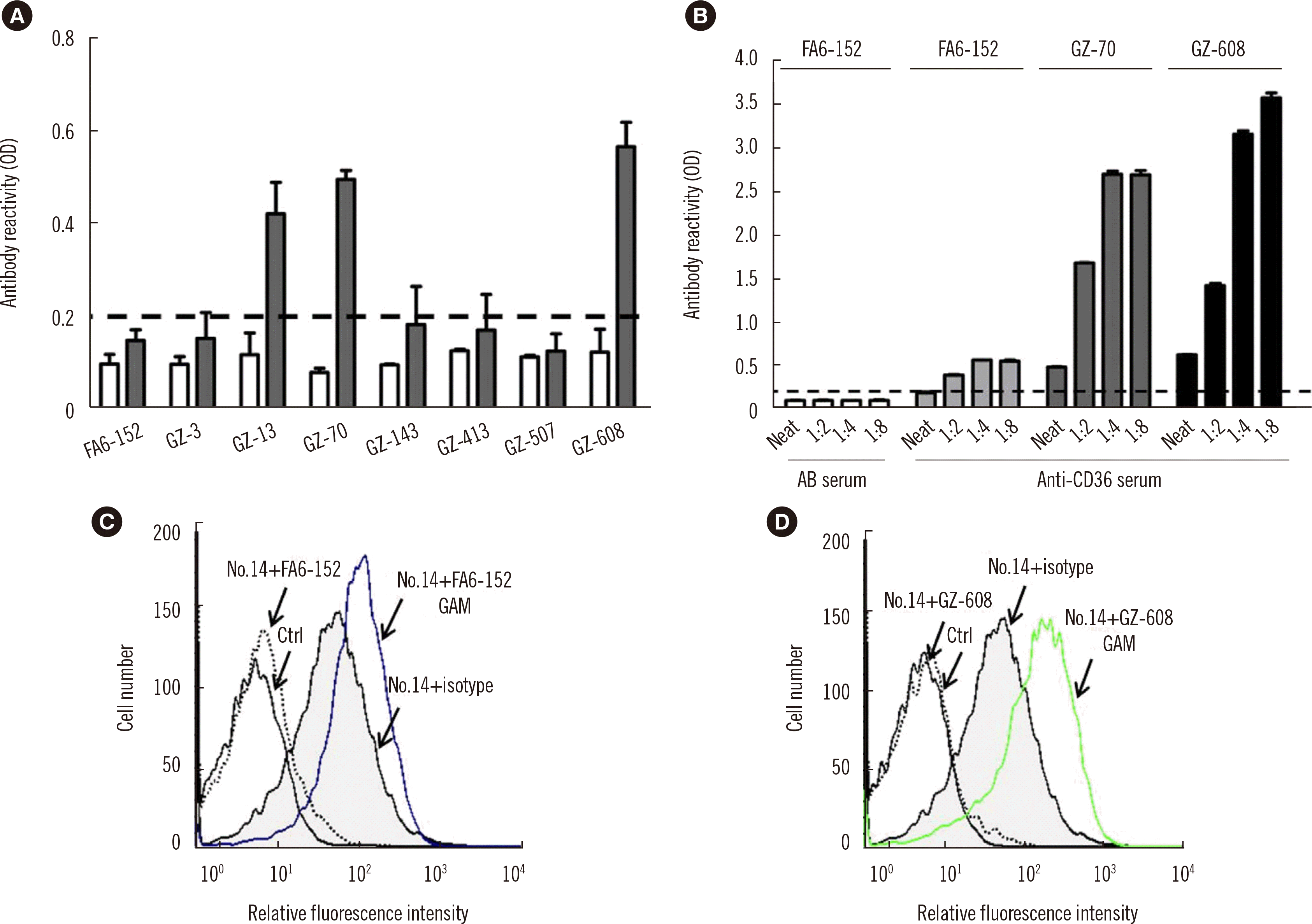 | Fig. 2
Characteristics of anti-CD36 serum in MAIPA and binding assays using different capture mAbs. (A) Platelets were first incubated with anti-CD36 serum (serum number 8, gray columns) and AB serum (white columns) and then with eight anti-CD36 mAbs (20 μg/mL) as indicated and analyzed using the MAIPA assay. (B) Platelets were incubated with anti-CD36 serum at different dilutions (neat, 1:2, 1:4, and 1:8) and the anti-CD36 mAbs FA6-152, GZ-70, and GZ-608 (20 μg/mL), as indicated. The cut-off for each assay was determined by analyzing AB sera from healthy blood donors (N=8, white columns). The reaction was considered positive when the result was >0.200 (cut-off; mean value±3 SDs; N=8; dotted line). (C, D) Effects of the mAbs FA6-152 and GZ-608 on the binding of serum number 14 to CD36+ platelets as determined using flow cytometry. “Isotype” stands for mouse IgG1, “Ctrl” for AB serum+mAb, and “GAM” for fluorescence-labeled goat anti-mouse IgG antibody.
Abbreviations: OD, optical density; MAIPA, monoclonal antibody immobilization of platelet antigens; mAbs, monoclonal antibodies.

|
Table 1
Reactivity of anti-CD36 sera in solid-phase ELISA (PakPlus) and/or MAIPA assays using three mAbs against CD36 (FA6-152, GZ-608, and GZ-70)
|
Anti-CD36 serum number |
Age (yr)/ sex |
Diagnosis |
Optical density |
|
|
PakPlus |
mAb FA6-152 |
mAb GZ-608 |
mAb GZ-70 |
|
1 |
44/M |
Healthy donor |
0.989 |
0.293 |
0.667 |
0.516 |
|
2 |
50/F |
Healthy donor |
1.138 |
0.438 |
1.003 |
0.733 |
|
3 |
50/M |
Hypercholesterolemia |
0.663 |
0.168
|
0.655 |
0.540 |
|
4 |
69/F |
Anemia |
0.791 |
0.200
|
0.797 |
0.341 |
|
5 |
41/M |
Thoracic aortic aneurysm |
0.569 |
0.145
|
0.255 |
0.290 |
|
6 |
22/M |
T-lymphoblastic cell tumor |
1.812 |
1.615 |
3.143 |
2.654 |
|
7 |
35/F |
Myelodysplastic syndromes |
0.923 |
0.431 |
0.754 |
0.696 |
|
8 |
35/F |
FNAIT (abortion) |
0.835 |
0.189
|
0.624 |
0.481 |
|
9 |
30/F |
FNAIT (abortion) |
1.551 |
0.020
|
0.550 |
0.440 |
|
10 |
36/F |
FNAIT (thrombocytopenia) |
0.990 |
0.200
|
0.355 |
0.375 |
|
11 |
46/M |
Alcoholic cirrhosis |
2.292 |
0.340 |
0.384 |
0.367 |
|
12 |
44/F |
Acute leukemia |
0.745 |
0.185
|
0.582 |
0.411 |
|
13 |
35/F |
FNAIT (thrombocytopenia) |
0.257 |
0.289 |
0.543 |
0.633 |
|
14 |
56/M |
Myelodysplastic syndrome |
0.827 |
0.199
|
0.143
|
0.158
|
|
|
Cut-off*
|
> 0.160 |
> 0.200 |
> 0.200 |
> 0.200 |
|
|
|
14/14 (100.0%) |
6/14 (42.9%) |
13/14 (92.9%) |
13/14 (92.9%) |

The three reactive mAbs, GZ-70, GZ-608, and FA6-152, were used to analyze our serum cohort (N=14). Anti-CD36 Abs were detected in only 6/14 (42.9%) of the sera tested with FA6-152 mAb. In contrast, anti-CD36 Abs were detected in 13/14 (92.9%) sera using GZ-70 or GZ-608 mAb (
Table 1). An additional serum cohort comprising four sera from patients with FNAIT (N=1) and TRALI (N=3) was tested using our assays [
10,
17]; all four sera showed positive reactions (Supplemental Data Table S1).
To exclude other factors that may decrease MAIPA assay sensitivity, such as the prozone effect (i.e., an unfavorable ratio between human and mouse anti-CD36 Abs), serum numbers 8, 11, and 12 (
Table 1) were retested at different dilutions (neat, 1:2, 1:4, 1:8). A weak positive reaction was detected in serum number 8 with FA6-152 mAb after dilutions. Significantly increased reactivity was found with GZ-70 and GZ-608 mAbs after dilutions (
Fig. 2B). In contrast, decreased reactivity was detected with serum numbers 11 and 12 (data not shown). These results showed that not only the choice of mAb but also dilution of the test serum can increase MAIPA assay sensitivity.
Interestingly, a negative result was obtained for serum number 14 (
Table 1). To clarify this phenomenon, the direct inhibitory effect of anti-CD36 mAbs on anti-CD36 Ab binding to platelets was analyzed. Fifty microliters of CD36-positive platelets (40×106 cells) was incubated with 25 µL of serum (at 1:16 dilution) at 37°C for 30 minutes and then with 25 µL of anti-CD36 mAb (20 µg/mL). After washing with EDTA/phosphate-buffered saline (PBS) (Dulbecco’s PBS containing 10 mM EDTA, pH 7.0–7.2; Gibco BRL, Waltham, MA, USA), 50 µL of fluorescein-conjugated goat anti-human IgG or goat anti-mouse IgG (Jackson, West Grove, PA, USA) was added, and the samples were incubated for another 30 minutes. After washing, bound platelets were suspended in 0.5 mL of EDTA/PBS and analyzed by flow cytometry. The binding of anti-CD36 Abs in serum number 14 to platelets was disrupted upon incubation with mAb FA6-152 (
Fig. 2C); similar results were found with GZ-608 (
Fig. 2D) and mAb GZ-70 (data not shown) mAbs. These results indicated that serum number 14 contained lower-avidity anti-CD36 Abs that can be displaced by higher-avidity anti-CD36 mAbs that bind to the same or similar epitopes; these results were particularly obtained due to a rigorous MAIPA washing procedure.
Most anti-CD36 mAbs, including FA6-152, OKM5, and 10/5, bind to epitopes within the domain that comprises amino acid residues 155–183 and prevents anti-CD36 Abs in serum samples from binding [
15,
18]. To avoid this, the use of a MAIPA panel containing different mAbs is recommended, but which mAbs should be included in the panel has not been defined.
MAbs against human CD36 have been produced through the immunization of mice with human fetal erythrocytes (FA6-152), human monocytes (OKM-5), or human platelets (10/5) [
15,
18-
20]. We generated mAbs against mouse CD36 through immunization of CD36-deficient mice with mouse CD36-transfected HEK293T cells and selected those mAbs that cross-reacted with human CD36. Through this strategy, we presumably obtained mAbs that do not recognize the highly immunogenic region(s) of human CD36 (residues 155–183), which harbor the major epitopes of human anti-CD36 Abs. Hence, in contrast to FA6-152, some of our mAbs (e.g., GZ-70 and GZ-608) had a significantly higher MAIPA reactivity (42.9% vs. 92.9%), indicating that these capture Abs did not compete with anti-CD36 Abs present in the sera. The anti-CD36 mAb 13/10 binds to another domain that spans residues 30–76. This unique mAb did not inhibit the binding of human anti-CD36 Abs [
15,
18].
Although the existence of an immunodominant region in CD36 (residues 155–183) has been reported, little is known about the exact location and diversity of epitopes recognized by polyclonal anti-CD36 Abs. That three mAbs showed differential inhibition suggests that at least two antigenic determinants exist. Additional information about the structure of the exact binding site of different anti-CD36 mAbs may help to prevent competitive inhibition.
The combination of antigen phenotyping (e.g., by flow cytometry) and Ab detection (e.g., by PakPlus) is reliable, convenient, and sufficient for the detection of isoantibodies against CD36. Given the existence of numerous naturally occurring single-nucleotide polymorphisms (SNPs) and the immunogenicity of CD36, individuals with normal (wild-type) CD36 expression may develop alloantibodies against CD36 due to alloimmunization with donor platelets carrying certain SNPs in certain cases, such as PTR and FNAIT. The existence of such cases can only be identified by crossmatch analysis (e.g., recipient serum vs. donor platelets), especially through glycoprotein specific immunoassay use.
In summary, improving MAIPA assays by applying selected noncompetitive mAb(s) will facilitate the screening of anti-CD36 Abs and the identification of new platelet antigens/Abs in patients with immune-mediated thrombocytopenia and other related disorders. However, further study in a larger cohort is necessary to evaluate the sensitivity of the MAIPA assay for the detection of anti-CD36 Abs.
Go to :







 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download