Dear Editor,
ETV6–RUNX1 (previously TEL–AML1) is the most common fusion gene associated with childhood ALL, which occurs in approximately 25% of pediatric ALL patients receiving diagnoses between the ages of 2–10 years and is associated with favorable prognoses [1, 2]. ETV6 rearrangement contributes to leukemogenesis in lymphoid neoplasms but is not well described in myeloid leukemia, and very few cases have been reported [3-6]. We report the first case of AML with the ETV6–RUNX1 fusion gene. The Institutional Review Board of Chonnam National University Hwasun Hospital, Hwasun, Korea, approved this study (CNUHH-2022-013) and waived the need for informed consent.
In January 2021, a 66-year-old man presented with newly developed thrombocytopenia and underlying dyslipidemia. Complete blood count showed leukocytosis (13.3×109/L) and thrombocytopenia (82×109/L). Peripheral blood smear comprised ~78% abnormal-appearing monocytes and nucleated RBCs. The bone marrow (BM) aspirate showed 63%, 7%, and 7% monoblasts, promonocytes, and monocytes, respectively (Fig. 1A). Myeloperoxidase (MPO) and non-specific esterase were positive in cytochemical staining (Fig. 1B, C). Flow cytometry analysis (Navios, Beckman Coulter, Miami, FL, USA) with BM cells identified two cell populations: the blast population showed a positive reaction in anti-CD33, anti-CD64, anti-CD117, and anti-HLA-DR fractions, whereas the monocyte population was positive for anti-CD13, anti-CD14, anti-CD33, anti-CD64, anti-HLA-DR, and anti-MPO. Both populations were negative for the lymphoid markers CD2, CD5, CD7, CD10, CD19, CD20, and CD22; only cytoplasmic CD3 showed aberrant dim expression in the monocyte population (~11.3%).
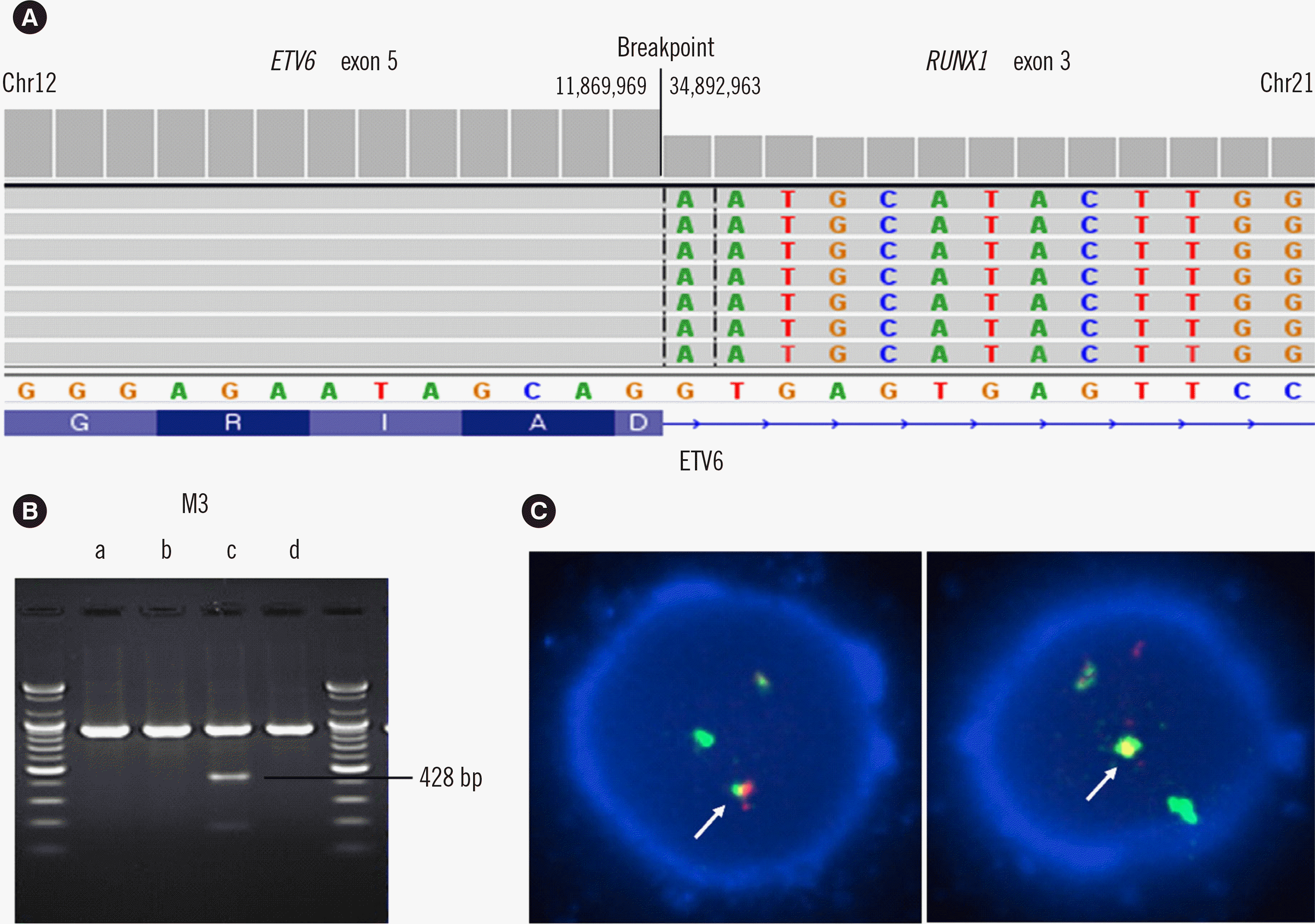
Conventional cytogenetic analysis demonstrated a normal karyotype. Based on morphologic and immunophenotypic results, a diagnosis of AML-M5a was made. The ETV6–RUNX1 fusion gene was detected using next-generation sequencing (NGS) RNA panel (KBB-RNAseq NGS-Leukemia-PHB; KBlueBio Inc., Hwasun, Korea) and confirmed with multiplex reverse transcription (RT)-PCR (HemaVision Kit; DNA Technology, Aarhus, Denmark). Other genetic variants, including SETBP1, JAK2, PHF6, and STAT3, were detected with an NGS DNA sequencing panel (HEMEaccu Test DNA; Ngenebio, Seoul, Korea); these variants are known to be related to myeloid rather than lymphoid neoplasms (Fig. 2A, B) [7].
Twenty-six days after induction chemotherapy, follow-up BM biopsy showed 49% monoblasts and 19% promonocytes. The samples were positive (10/200) for the reciprocal translocation t(12;21)(p13;q22), as demonstrated by FISH with a t(12;21) probe (Vysis, Abbott Molecular, Des Plaines, IL, USA) (Fig. 2C). Repeated BM biopsy 47 days after reinduction chemotherapy showed no monoblasts or promonocytes, and the sample was negative for t(12;21)(p13;q22) in FISH analysis. Consolidation chemotherapy was continued for five days. Despite long-term additional treatment because of neutropenic fever, neurologic symptoms became worse, accompanied by a confused mental state without clear reason. The patient was discharged without curative hope on day 226 after the diagnosis.
RUNX1 encodes a transcription factor (AML1) that controls the expression of its target genes involved in hematopoietic differentiation, and its functional disruption is a major causative factor of leukemia development [8, 9]. The RUNX1–RUNX1T1 fusion product in AML contains only the N-terminal portion of AML1, whereas the ETV6–RUNX1 fusion product includes almost the entire AML1 sequence, containing both the Runt homology domain and C-terminal transactivating and repressor domains [4, 9]. ETV6–RUNX1 translocation results in the production of a fusion protein that likely acts in a dominant-negative fashion to interfere with the normal function of RUNX1 and is considered an early lesion in B-cell leukemogenesis [2, 9]. A two-hit model of ETV6–RUNX1 leukemogenesis is assumed: ETV6–RUNX1 expression and a second genetic or environmental hit must occur at the hematopoietic progenitor stage for leukemic transformation [10]. ETV6–RUNX1 fusion is considered to be absent in leukemias of myeloid lineage because the fusion protein severely interferes with myeloid development and enforced expression of ETV6–RUNX1 in myeloid cells leads to apoptosis, possibly by interfering with autoregulatory loops essential for the maintenance of myeloid identity [3]. Presence of the ETV6–RUNX1 fusion gene in AML patients is very rare, and we found no report describing this association. The ETV6–RUNX1 fusion is considered to occur in adult patients, resulting in leukemogenesis to myeloid leukemia caused by uncommon second hits, representing a distinct mechanism from pediatric ALL.
In this case, the ETV6–RUNX1 fusion gene was detected at diagnosis and follow-up status but disappeared at morphological remission status. We tentatively conclude that the ETV6–RUNX1 fusion played an important role in the pathogenesis of AML in this case; however, the mechanism is unclear, which is the study’s limitation. Although further studies are needed to determine the leukemogenesis of the ETV6–RUNX1 fusion gene in AML, our case demonstrates the possibility of this fusion gene in adult de novo AML.
Notes
AUTHOR CONTRIBUTIONS
Jeon SJ and Park JH conceived and designed the study, collected and analyzed the data, and drafted the manuscript. Park JH and Shin MG revised the manuscript. Lim HJ, Won EJ, Choi HW, Choi HJ, Kee SJ, Kim SH, and Shin JH provided valuable comments and recommendations. All authors have accepted their responsibility for the entire content of this manuscript and approved the submission.
REFERENCES
1. Linka Y, Ginzel S, Krüger M, Novosel A, Gombert M, Kremmer E, et al. 2013; The impact of TEL-AML1 (ETV6-RUNX1) expression in precursor B cells and implications for leukaemia using three different genome-wide screening methods. Blood Cancer J. 3:e151. DOI: 10.1038/bcj.2013.48. PMID: 24121163. PMCID: PMC3816209.

2. Swerdlow SH, Campo E, editors. 2008. WHO classification of tumours of haematopoietic and lymphoid tissues. 5th ed. International Agency for Research on Cancer;Lyon: p. 250.
3. Bohlander SK. 2005; ETV6: a versatile player in leukemogenesis. Semin Cancer Biol. 15:162–74. DOI: 10.1016/j.semcancer.2005.01.008. PMID: 15826831.

4. Fischer M, Schwieger M, Horn S, Niebuhr B, Ford A, Roscher S, et al. 2005; Defining the oncogenic function of the TEL/AML1 (ETV6/RUNX1) fusion protein in a mouse model. Oncogene. 24:7579–91. DOI: 10.1038/sj.onc.1208931. PMID: 16044150.

5. Lee JJ, Cho H, Park KS, Kim YJ, Cho SY, Han JJ, et al. 2021; First case report of acute myeloid leukemia with MN1-ETV6 rearrangement at relapse. Lab Med Online. 11:145–8. DOI: 10.47429/lmo.2021.11.2.145.

6. Kim KH, Kim MJ, Ahn JY, Park PW, Seo YH, Jeong JH. 2016; Acute myeloid leukemia with t(4;12)(q12;p13): report of 2 cases. Blood Res. 51:133–7. DOI: 10.5045/br.2016.51.2.133. PMID: 27382559. PMCID: PMC4931932.

7. Higgins A, Shah MV. 2020; Genetic and genomic landscape of secondary and therapy-related acute myeloid leukemia. Genes (Basel). 11:749. DOI: 10.3390/genes11070749. PMID: 32640569. PMCID: PMC7397259.

8. Deltcheva E, Nimmo R. 2017; RUNX transcription factors at the interface of stem cells and cancer. Biochem J. 474:1755–68. DOI: 10.1042/BCJ20160632. PMID: 28490659.

9. Sood R, Kamikubo Y, Liu P. Role of RUNX1 in hematological malignancies. Blood. 2017; 129:2070–82. DOI: 10.1182/blood-2016-10-687830. PMID: 28179279. PMCID: PMC5391618.

10. Rodríguez-Hernández G, Casado-García A, Isidro-Hernández M, Picard D, Raboso-Gallego J, Alemán-Arteaga S, et al. 2021; The second oncogenic hit determines the cell fate of ETV6-RUNX1 positive leukemia. Front Cell Dev Biol. 9:704591. DOI: 10.3389/fcell.2021.704591. PMID: 34336858. PMCID: PMC8320889.

Fig. 1
Results of bone marrow aspirate analysis. Blasts have a moderate nuclear-cytoplasmic ratio, round nuclei with finely dispersed chromatin, and a few distinct nucleoli, some with tiny vacuoles. (A) Wright-Giemsa stain, ×1,000. (B) Myeloperoxidase stain, ×1,000. (C) Non-specific esterase stain, ×1,000.
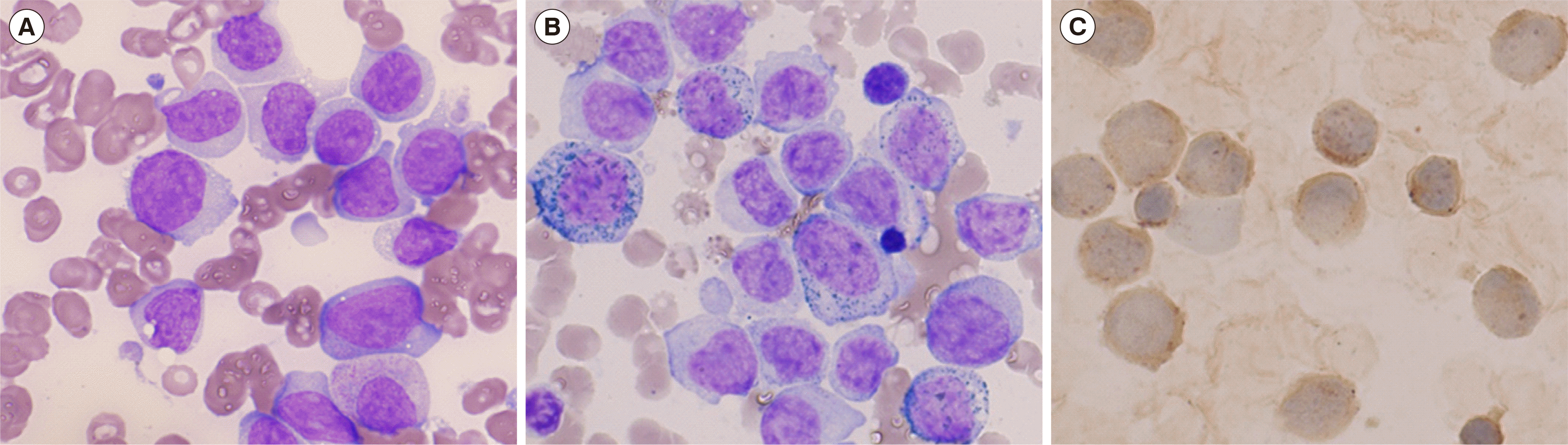
Fig. 2
ETV6-RUNX1 fusion finding with various methods. (A) Integrative genomics viewer image showing the ETV6–RUNX1 breakpoints. Fourteen fusion reads were found within 722 ETV6 reference reads. (B) Reverse transcription-PCR results showing the fusion between ETV6 exon 5 and RUNX1 exon 3 (428 bp). (C) FISH on the interphase nucleus with red signals (ETV6), green signals (RUNX1), and yellow signals (ETV6–RUNX1 fusion site, arrow).




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download