1. Manoharan A, Manchanda V, Balasubramanian S, Lalwani S, Modak M, Bai S, et al. 2017; Invasive pneumococcal disease in children aged younger than 5 years in India: a surveillance study. Lancet Infect Dis. 17:305–12. DOI:
10.1016/S1473-3099(16)30466-2. PMID:
27956163.

2. Park DC, Kim SH, Yong D, Suh IB, Kim YR, Yi J, et al. 2019; Serotype distribution and antimicrobial resistance of invasive and noninvasive
Streptococcus pneumoniae isolates in Korea between 2014 and 2016. Ann Lab Med. 39:537–44. DOI:
10.3343/alm.2019.39.6.537. PMID:
31240881. PMCID:
PMC6660335.

3. Croney CM, Nahm MH, Juhn SK, Briles DE, Crain MJ. 2013; Invasive and noninvasive
Streptococcus pneumoniae capsule and surface protein diversity following the use of a conjugate vaccine. Clin Vaccine Immunol. 20:1711–8. DOI:
10.1128/CVI.00381-13. PMID:
24006139. PMCID:
PMC3837785.

4. Diawara I, Zerouali K, Katfy K, Zaki B, Belabbes H, Najib J, et al. 2015; Invasive pneumococcal disease among children younger than 5 years of age before and after introduction of pneumococcal conjugate vaccine in Casablanca, Morocco. Int J Infect Dis. 40:95–101. DOI:
10.1016/j.ijid.2015.09.019. PMID:
26434380.

5. Moreno J, Duarte C, Cassiolato AP, Chacón GC, Alarcon P, Sánchez J, et al. 2020; Molecular characterization of Latin American invasive
Streptococcus pneumoniae serotype 19A isolates. Vaccine. 38:3524–30. DOI:
10.1016/j.vaccine.2020.03.030. PMID:
32204942.

6. Kim CJ, Song JS, Choi SJ, Song KH, Choe PG, Park WB, et al. 2016; Serotype distribution and antimicrobial susceptibilities of invasive
Streptococcus pneumoniae isolates from adults in Korea from 1997 to 2012. J Korean Med Sci. 31:715–23. DOI:
10.3346/jkms.2016.31.5.715. PMID:
27134492. PMCID:
PMC4835596.

7. Kim SH, Chung DR, Song JH, Baek JY, Thamlikitkul V, Wang H, et al. 2020; Changes in serotype distribution and antimicrobial resistance of
Streptococcus pneumoniae isolates from adult patients in Asia: emergence of drug-resistant non-vaccine serotypes. Vaccine. 38:6065–73. DOI:
10.1016/j.vaccine.2019.09.065. PMID:
31590932.

8. Park D, Kim SH, Bae IK, Kim NY, Kook JK, Park YH, et al. 2019; Evaluation of modified sequential multiplex PCR for
Streptococcus pneumoniae serotyping. Jpn J Infect Dis. 72:224–7. DOI:
10.7883/yoken.JJID.2018.422. PMID:
30814459.

9. Habib M, Porter BD, Satzke C. Capsular serotyping of
Streptococcus pneumoniae using the Quellung reaction. J Vis Exp. 2014; (84):e51208. DOI:
10.3791/51208. PMID:
24637727. PMCID:
PMC4131683.

10. Steenhoff AP, Shah SS, Ratner AJ, Patil SM, McGowan KL. 2006; Emergence of vaccine-related pneumococcal serotypes as a cause of bacteremia. Clin Infect Dis. 42:907–14. DOI:
10.1086/500941. PMID:
16511752.

11. CLSI. 2022. Performance standards for antimicrobial susceptibility testing. 32nd ed. CLSI M100. Clinical and Laboratory Standards Institute;Wayne, PA: DOI:
10.1086/500941.
12. Golden AR, Rosenthal M, Fultz B, Nichol KA, Adam HJ, Gilmour MW, et al. 2015; Characterization of MDR and XDR
Streptococcus pneumoniae in Canada, 2007-13. J Antimicrob Chemother. 70:2199–202. DOI:
10.1093/jac/dkv107. PMID:
25921512.
13. Enright MC, Spratt BG. 1998; A multilocus sequence typing scheme for
Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology (Reading). 144:3049–60. DOI:
10.1099/00221287-144-11-3049. PMID:
9846740.
15. Wu CJ, Lai JF, Huang IW, Shiau YR, Wang HY, Lauderdale TL. 2020; Serotype distribution and antimicrobial susceptibility of
Streptococcus pneumoniae in pre- and post- PCV7/13 eras, Taiwan, 2002-2018. Front Microbiol. 11:557404. DOI:
10.3389/fmicb.2020.557404. PMID:
33193140. PMCID:
PMC7642986.
16. Harboe ZB, Dalby T, Weinberger DM, Benfield T, Mølbak K, Slotved HC, et al. 2014; Impact of 13-valent pneumococcal conjugate vaccination in invasive pneumococcal disease incidence and mortality. Clin Infect Dis. 59:1066–73. DOI:
10.1093/cid/ciu524. PMID:
25034421.

17. Hink RK, Adam HJ, Golden AR, Baxter M, Martin I, Nichol KA, et al. 2021; Comparison of PCV-10 and PCV-13 vaccine coverage for invasive pneumococcal isolates obtained across Canadian geographic regions, SAVE 2011 to 2017. Diagn Microbiol Infect Dis. 99:115282. DOI:
10.1016/j.diagmicrobio.2020.115282. PMID:
33341491.

18. Suaya JA, Mendes RE, Sings HL, Arguedas A, Reinert RR, Jodar L, et al. 2020;
Streptococcus pneumoniae serotype distribution and antimicrobial nonsusceptibility trends among adults with pneumonia in the United States, 2009-2017. J Infect. 81:557–66. DOI:
10.1016/j.jinf.2020.07.035. PMID:
32739491.
19. Ludwig G, Garcia-Garcia S, Lanaspa M, Ciruela P, Esteva C, Fernandez de Sevilla M, et al. 2020; Serotype and clonal distribution dynamics of invasive pneumococcal strains after PCV13 introduction (2011-2016): Surveillance data from 23 sites in Catalonia, Spain. PLoS One. 15:e0228612. DOI:
10.1371/journal.pone.0228612. PMID:
32027715. PMCID:
PMC7004304.

20. Yanagihara K, Kosai K, Mikamo H, Mukae H, Takesue Y, Abe M, et al. 2021; Serotype distribution and antimicrobial susceptibility of
Streptococcus pneumoniae associated with invasive pneumococcal disease among adults in Japan. Int J Infect Dis. 102:260–8. DOI:
10.1016/j.ijid.2020.10.017. PMID:
33065297.
21. Lee S, Bae S, Lee KJ, Yu JY, Kang Y. 2013; Changes in serotype prevalence and antimicrobial resistance among invasive
Streptococcus pneumoniae isolates in Korea, 1996-2008. J Med Microbiol. 62:1204–10. DOI:
10.1099/jmm.0.058164-0. PMID:
23657529.
22. Park M, Kim HS, Shin KS, Kim HS, Park JY, Song W, et al. 2014; Changes in the incidence of
Streptococcus pneumoniae bacteremia and its serotypes over 10 years in one hospital in South Korea. Vaccine. 32:6403–7. DOI:
10.1016/j.vaccine.2014.09.062. PMID:
25305566.

23. Gaviria-Agudelo CL, Jordan-Villegas A, Garcia C, McCracken GH Jr. 2017; The effect of 13-valent pneumococcal conjugate vaccine on the serotype distribution and antibiotic resistance profiles in children with invasive pneumococcal disease. J Pediatric Infect Dis Soc. 6:253–9. DOI:
10.1093/jpids/piw005. PMID:
26907814. PMCID:
PMC7107452.

24. Levy C, Varon E, Ouldali N, Béchet S, Bonacorsi S, Cohen R. 2020; Changes in invasive pneumococcal disease spectrum after 13-valent pneumococcal conjugate vaccine implementation. Clin Infect Dis. 70:446–54. DOI:
10.1093/cid/ciz221. PMID:
30869777.

25. Nakano S, Fujisawa T, Ito Y, Chang B, Matsumura Y, Yamamoto M, et al. 2020; Nationwide surveillance of paediatric invasive and non-invasive pneumococcal disease in Japan after the introduction of the 13-valent conjugated vaccine, 2015-2017. Vaccine. 38:1818–24. DOI:
10.1016/j.vaccine.2019.12.022. PMID:
31882246.

26. Desmet S, Wouters I, Heirstraeten LV, Beutels P, Van Damme P, Malhotra-Kumar S, et al. 2021; In-depth analysis of pneumococcal serotypes in Belgian children (2015-2018): diversity, invasive disease potential, and antimicrobial susceptibility in carriage and disease. Vaccine. 39:372–9. DOI:
10.1016/j.vaccine.2020.11.044. PMID:
33308889.

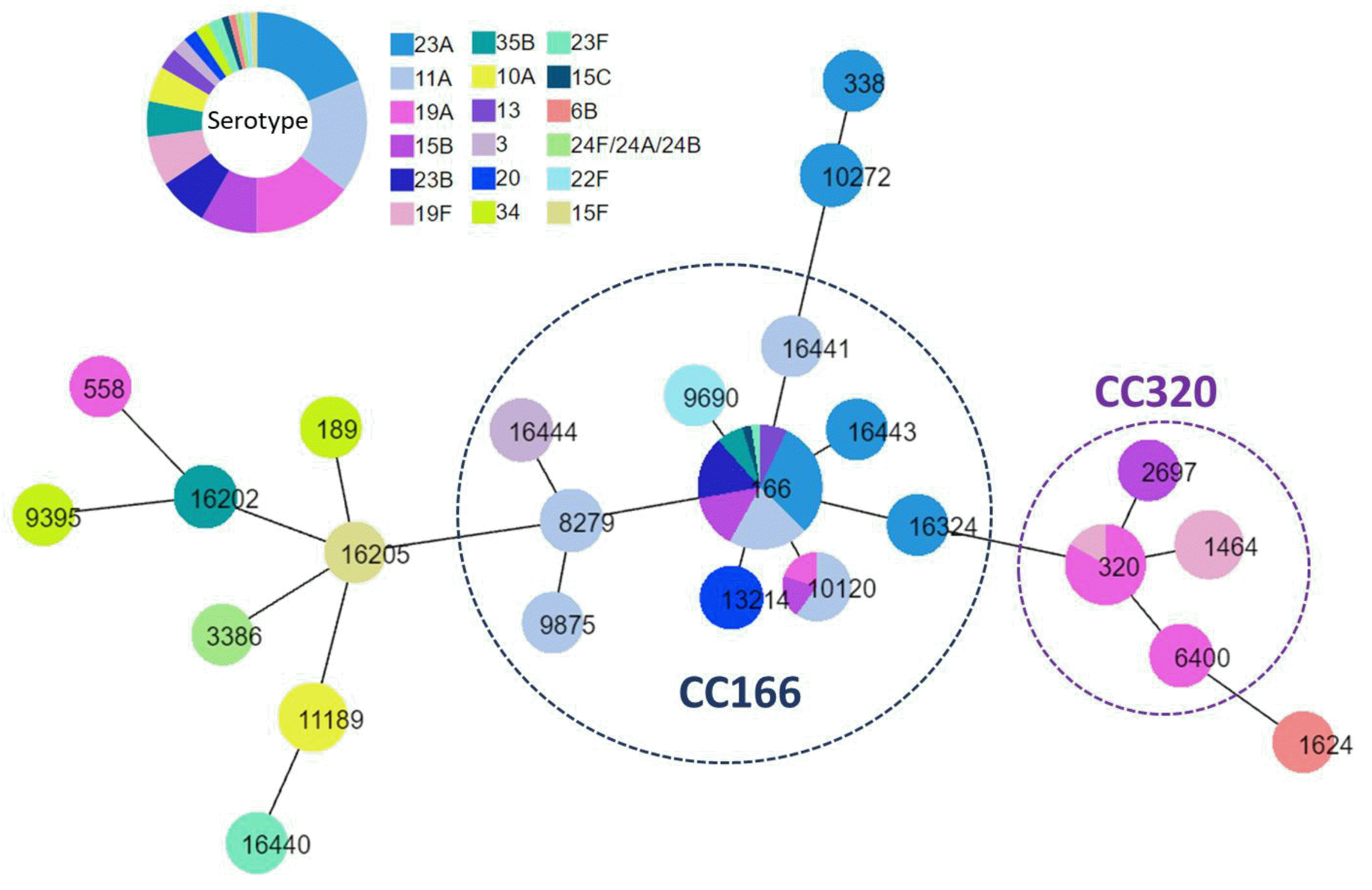
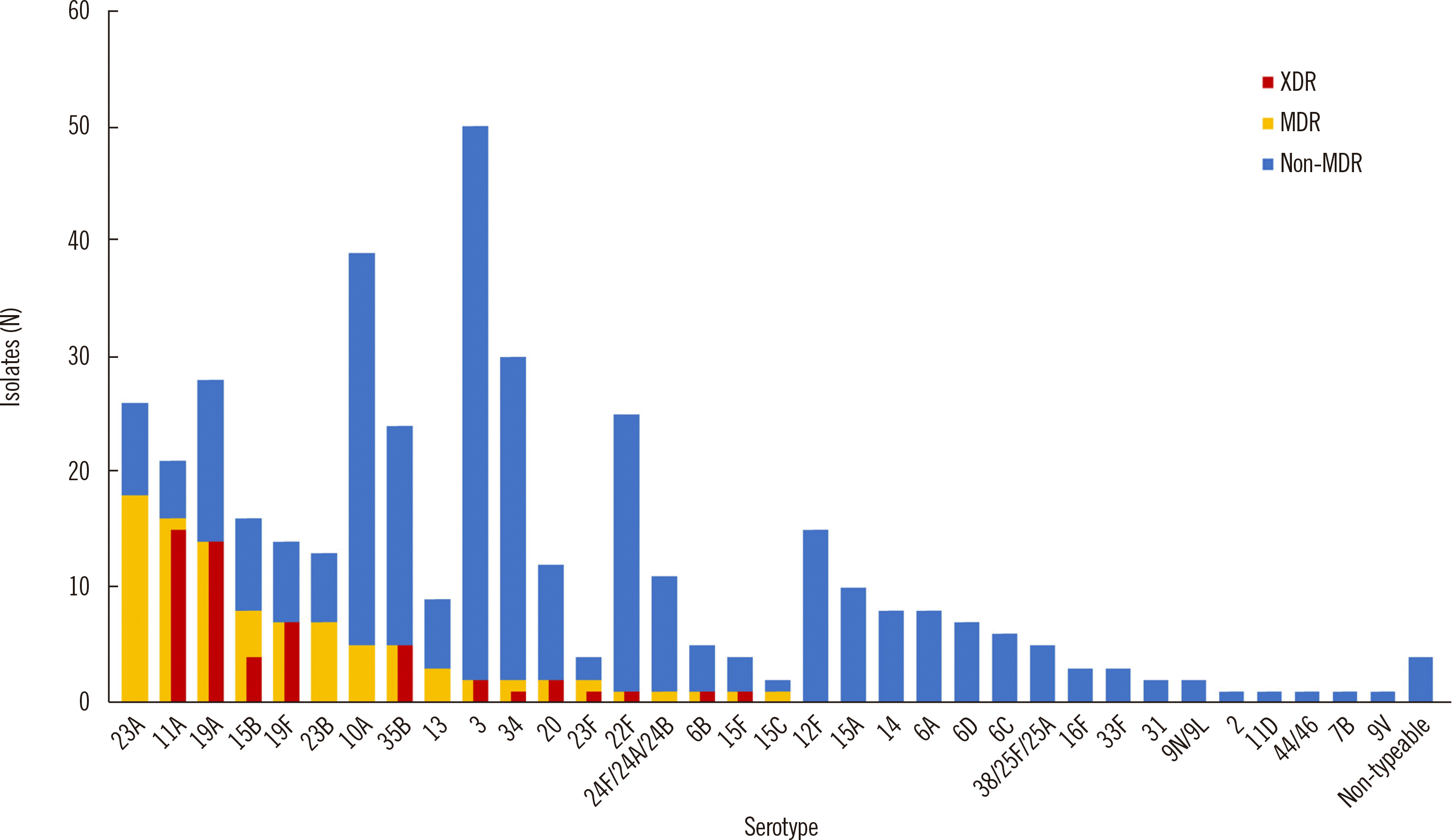
27. Yun KW, Rhie K, Kang JH, Kim KH, Ahn JG, Kim YJ, et al. 2021; Emergence of serotype 10A-ST11189 among pediatric invasive pneumococcal diseases, South Korea, 2014-2019. Vaccine. 39:5787–93. DOI:
10.1016/j.vaccine.2021.08.072. PMID:
34465475.

28. Ubukata K, Takata M, Morozumi M, Chiba N, Wajima T, Hanada S, et al. 2018; Effects of pneumococcal conjugate vaccine on genotypic penicillin resistance and serotype changes, Japan, 2010-2017. Emerg Infect Dis. 24:2010–20. DOI:
10.3201/eid2411.180326. PMID:
30334707. PMCID:
PMC6200004.

29. Imöhl M, Reinert RR, Ocklenburg C, van der Linden M. 2010; Association of serotypes of
Streptococcus pneumoniae with age in invasive pneumococcal disease. J Clin Microbiol. 48:1291–6. DOI:
10.1128/JCM.01937-09. PMID:
20107087. PMCID:
PMC2849605.
30. Olarte L, Kaplan SL, Barson WJ, Romero JR, Lin PL, Tan TQ, et al. 2017; Emergence of multidrug-resistant pneumococcal serotype 35B among children in the United States. J Clin Microbiol. 55:724–34. DOI:
10.1128/JCM.01778-16. PMID:
27847379. PMCID:
PMC5328440.

31. Lo SW, Gladstone RA, van Tonder AJ, Lees JA, du Plessis M, Benisty R, et al. 2019; Pneumococcal lineages associated with serotype replacement and antibiotic resistance in childhood invasive pneumococcal disease in the post-PCV13 era: an international whole-genome sequencing study. Lancet Infect Dis. 19:759–69. DOI:
10.1016/S1473-3099(19)30297-X. PMID:
31196809. PMCID:
PMC7641901.
32. Suzuki S, Osato R, Wajima T, Hasebe T, Ishikawa H, Mitsumori H, et al. 2019; Impact of the introduction of a 13-valent pneumococcal vaccine on pneumococcal serotypes in non-invasive isolates from 2007 to 2016 at a teaching hospital in Japan. J Med Microbiol. 68:903–9. DOI:
10.1099/jmm.0.000992. PMID:
31090535.

33. Zhao C, Xie Y, Zhang F, Wang Z, Yang S, Wang Q, et al. 2020; Investigation of antibiotic resistance, serotype distribution, and genetic characteristics of 164 invasive
Streptococcus pneumoniae from North China between April 2016 and October 2017. Infect Drug Resist. 13:2117–28. DOI:
10.2147/IDR.S256663. PMID:
32753907. PMCID:
PMC7342493.
34. Moore CE, Paul J, Foster D, Mahar SA, Griffiths D, Knox K, et al. 2014; Reduction of invasive pneumococcal disease 3 years after the introduction of the 13-valent conjugate vaccine in the Oxfordshire region of England. J Infect Dis. 210:1001–11. DOI:
10.1093/infdis/jiu213. PMID:
24719477.

35. Tsai YT, Lee YL, Lu MC, Shao PL, Lu PL, Cheng SH, et al. 2022; Nationwide surveillance of antimicrobial resistance in invasive isolates of
Streptococcus pneumoniae in Taiwan from 2017 to 2019. J Microbiol Immunol Infect. 55:215–24. DOI:
10.1016/j.jmii.2021.05.008. PMID:
34219043.
36. Bao Y, Wang Q, Yao K, Xie G, Gao W, Huang L, et al. 2019; The changing phenotypes and genotypes of invasive pneumococcal isolates from children in Shenzhen during 2013-2017. Vaccine. 37:7248–55. DOI:
10.1016/j.vaccine.2019.09.069. PMID:
31635974.

37. Wang Q, Shi W, Li Y, Gao W, Yuan L, Dong F, et al. 2020; Serotype distribution of
Streptococcus pneumoniae isolated from children hospitalized in Beijing Children's Hospital (2013-2019). Vaccine. 38:7858–64. DOI:
10.1016/j.vaccine.2020.10.005. PMID:
33164807.

38. Baek JY, Kim SH, Kang CI, Chung DR, Peck KR, Ko KS, et al. 2016; Prevalence of antimicrobial resistant
Streptococcus pneumoniae serotype 11A isolates in Korea, during 2004-2013, due to the increase of multidrug-resistant clone, CC166. Infect Genet Evol. 38:122–5. DOI:
10.1016/j.meegid.2015.12.018. PMID:
26733441.
39. Park M, Kim HS, Kim HS, Park JY, Song W, Cho HC, et al. 2016; Novel levofloxacin-resistant multidrug-resistant
Streptococcus pneumoniae serotype 11A isolates, South Korea. Emerg Infect Dis. 22:1978–80. DOI:
10.3201/eid2211.151450. PMID:
27767906. PMCID:
PMC5088008.
40. Wang X, Cong Z, Huang W, Li C. 2020; Molecular characterization of
Streptococcus pneumoniae isolated from pediatric patients in Shanghai, China. Pediatr Pulmonol. 55:2135–41. DOI:
10.1002/ppul.24877. PMID:
32470194.

41. Yun KW, Choi EH, Lee HJ, Kang JH, Kim KH, Kim DS, et al. 2018; Genetic structures of invasive
Streptococcus pneumoniae isolates from Korean children obtained between 1995 and 2013. BMC Infect Dis. 18:268. DOI:
10.1186/s12879-018-3177-7. PMID:
29884115. PMCID:
PMC5994121.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download