Abstract
Background and Objectives
Difficulties often encountered in separating and purifying active muscle satellite cells (MSCs) from skeletal muscle tissues have limited the supply of cells for muscle therapy and artificial meat production. Here, we report an effective isolation protocol to economically and conveniently retrieve active MSCs from skeletal muscle tissues in mice.
Methods and Results
We optimized an enzyme-based tissue digestion protocol for isolating skeletal muscle-derived primary cell population having a large number of active MSCs and described a method of differential plating (DP) for improving purity of active MSCs from skeletal muscle-derived primary cell population. Then, the age of the mouse appropriate to the isolation of a large number of active MSCs was elucidated. The best isolation yield of active MSCs from mouse skeletal muscle tissues was induced by the application of DP method to the primary cell population harvested from skeletal muscle tissues of 2-week-old mice digested in 0.2% (w/v) collagenase type II for 30 min at 37℃ and then in 0.1% (w/v) pronase for 5 min at 37℃.
Conclusions
The protocol we developed not only facilitates the isolation of MSCs but also maximizes the retrieval of active MSCs. Our expectation is that this protocol will contribute to the development of original technologies essential for muscle therapy and artificial meat industrialization in the future.
Muscles regulate various physiological functions in the human body (1, 2). For example, skeletal muscles enable locomotion by regulating movements of the skeletal system based on contraction and relaxation of the muscles (2, 3). However, persistent repetition of muscle contraction and relaxation can induce progressive muscle damage (4). Furthermore, vigorous exercise and sudden physical movements or accidents can result in more severe damage (5, 6).
Fortunately, muscles are competent at regenerating injured areas and recovering their functional roles (7, 8). A mediator in the recovery of injured muscles is the muscle satellite cell (MSC) (8, 9). Myocytes differentiated from MSCs mobilize to damaged areas (10, 11) where they generate de novo myofibers (10, 12) that contribute to the regeneration of injured muscles (10-12). Because of their unique characteristics, MSCs can potentially be used as a cell-mediated therapeutic tool for curing muscle-related genetic disorders (13, 14), muscle atrophy arising from the deficiency of MSCs within aged muscles (2, 15), and damage or rupture of muscles caused by exercise or accidents (2, 16). In addition, MSCs could be a resource for producing artificial meat without additional animal sacrifice (17, 18). The successful production of artificial meat will play a pivotal role in reducing environmental pollution arising from livestock industries (19, 20) and fulfilling increased meat consumption without any additional pollution (18, 21). These applications demonstrate the importance and potential uses of MSCs.
MSCs constitute approximately 2∼5% of muscular tissue (22, 23). A variety of protocols for the isolation of MSCs from muscle tissue have recently been introduced, including pre-plating (24, 25), magnetic-activated cell sorting (MACS) (26, 27), and fluorescence-activated cell sorting (FACS) (28, 29). However, these protocols have one common limitation. The purification efficiency is too low to provide a sufficient amount of cells (30). Furthermore, the pre-plating method does not provide certainty regarding the time point when cell populations, including high-yield MSCs, can be collected (24, 25), and the isolation process using both the MACS and FACS methods is complicated and inconvenient (26-29). Therefore, a high-yield, simple, and convenient protocol for collecting MSCs from muscular tissues needs to be developed to facilitate basic or applied muscle-related research. Here, we present a new high-yield protocol to isolate active MSCs from mouse skeletal muscle tissues as well as analyze and compare the characteristics of active MSCs separated using different protocols.
Institute of Cancer Research (ICR) mice aged 1∼4 weeks purchased from DBL (Eumseong, Korea) were used as donors of MSCs. All animal handling, housing, and experimental procedures were performed in accordance with the Animal Care and Use Guidelines of Kangwon National University and approved by the Institutional Animal Care and Use Committee (IACUC) of Kangwon National University (IACUC approval no. KW-200421-1).
Mice were sacrificed via cervical dislocation. Skeletal muscle tissues collected from the hindlimbs were washed twice with antibiotic solution consisting of Dulbecco’s phosphate-buffered saline (DPBS; Welgene, Gyeongsan, Korea) supplemented with 1% (v/v) antibiotic-antimycotic (Welgene). The washed skeletal muscle tissues were cut into small pieces and washed once with the antibiotic solution. Subsequently, the small pieces of skeletal muscle tissues were enzymatically dissociated using three different methods (Fig. S1). Method 1 involved digestion in 0.2% (w/v) collagenase type II (Worthington Biochemical Corporation, Lakewood, NJ, USA) dissolved in high-glucose Dulbecco’s modified Eagle’s medium (HG-DMEM; Welgen) for 75 min at 37℃ (in a water bath). Method 2 involved digestion in 0.2% (w/v) collagenase type II and 0.125% (v/v) trypsin-EDTA (Welgene) dissolved in HG-DMEM for 40 min at 37℃ (in a water bath), and Method 3 involved digestion in 0.1% (w/v) pronase (Calbiochem, Darmstadt, Germany) dissolved in Earle’s balanced salt solution (Sigma-Aldrich, St. Louis, MO, USA) for 5 min at 37℃ (in a water bath) after incubation in 0.2% (w/v) collagenase type II dissolved in HG-DMEM for 30 min at 37℃ (in a water bath). Subsequently, the enzymes were inactivated by suspending the digested skeletal muscle tissues in HG-DMEM supplemented with 2% (v/v) heat-inactivated fetal bovine serum (FBS; Welgene). After centrifuging at 1500×g for 4 min, the supernatant was removed and red blood cells (RBCs) in the pellet were eliminated using RBC lysis buffer (Sigma-Aldrich) for 10 min at room temperature. The RBC-free cells were filtered using a 100-μm cell strainer (SPL, Pocheon, Korea), followed by filtration through a 70-μm cell strainer (SPL). The filtered cells were centrifuged for 5 min at 125×g, and the collected supernatant was additionally centrifuged for 5 min at 1000×g. The pellets were then retrieved, resuspended, and used for the following experiments.
Five times of 105 muscle-derived primary cells were seeded onto 35-mm culture dishes (SPL) and cultured in Ham’s F10 nutrient mixture (Gibco, Grand Island, NY, USA ) supplemented with 15% (v/v) FBS, 2.5 ng/ml basic fibroblast growth factor (Peprotech, Rocky Hill, NJ, USA), and 1% (v/v) antibiotic-antimycotic (herein referred to as proliferation medium). After culture for 24 h, floating (non-adherent) cells were collected and centrifuged at 1,500×g for 4 min. The pellets were then re-suspended in fresh proliferation medium and incubated in 35-mm culture dishes at 37℃ in a humidified atmosphere of 5% CO2 in air. After 72 h, adherent cells were retrieved via treatment with 0.25% trypsin-EDTA (Welgene), washed with fresh proliferation medium, and used for the following experiments. Details of the DP method are shown in Fig. S1.
Round coverslips (SPL) were coated with 0.01% (w/v) poly-L-lysine solution (Sigma-Aldrich) and left for 1 h at room temperature. After incubating the cells on the coated coverslip for 30 min, the attached cells were washed twice with DPBS and then fixed using 4% (v/v) formaldehyde solution (Junsei Chemical, Chuo-ku, Japan) at room temperature for 10 min. Fixed cells were washed twice with DPBS and incubated in blocking solution consisting of DPBS supplemented with 10% (v/v) heat-inactivated horse serum (Gibco), 2% (w/v) bovine serum albumin (Sigma-Aldrich), and 0.5% (v/v) Triton X-100 (Biopure, Cam-bridge, MA, USA) overnight at 4℃. The blocked cells were stained with fluorescence-unconjugated Pax7 primary antibody diluted in DPBS for 3 h at room temperature. The localization of the primary antibody was identified via incubation with Alexa Fluor 594-conjugated secondary antibody diluted in DPBS for 1 h at room temperature. Addi-tion-ally, the stained cells were incubated for 1 h at room temperature in Alexa Fluor 488-conjugated MyoD antibody diluted in DPBS. Table 1 shows the detailed information and dilutions of the antibodies used. The double-stained cells were washed three times with DPBS and counterstained with VECTASHIELD® Antifade mounting medium containing 4’,6-diamidino-2-phenylindole (Vector Labor-atories, Inc., Burlingame, CA, USA). Subs-equently, the triple-stained cells were observed under a fluorescence microscope (Olympus, Tokyo, Japan).
Numerical data were analyzed using Statistical Analysis System (SAS) software (SAS Institute, Cary, NC, USA). Comparative analysis among experimental groups was conducted using the least squares or Duncan’s method. The significance of the main effects was assessed using analysis of variance (ANOVA) with SAS software. p values less than 0.05 were regarded as indicating statistically significant differences.
To establish an optimal enzymatic digestion method for isolating primary cell populations containing a large number of active MSCs from mouse skeletal muscle tissues, we used three different methods to enzymatically digest muscle tissue (Fig. S1). Subsequently, the presence of active MSCs co-expressing Pax7 and MyoD (both active MSC-related markers) was evaluated. We identified Pax7+/MyoD+ MSCs within muscle-derived primary cell populations generated using each of the three digestion methods. Regardless of the method used, primary cell populations dissociated from skeletal muscle tissues contained Pax7+/MyoD+ MSCs (Fig. 1A). With no significant differences, all replicates derived using Method 3 (mean±SD, 21.27±3.67%) contained the highest proportion of Pax7+/MyoD+ MSCs in muscle-derived primary cell populations compared with those derived using Method 1 (mean±SD, 17.55±5.38%) and Method 2 (mean±SD, 14.45±2.28%) (Fig. 1B). These results indicate that muscle-derived primary cell populations containing large numbers of active MSCs can be harvested from skeletal muscle tissues via digestion in 0.1% (w/v) pronase for 5 min at 37℃ after incubation in 0.2% (w/v) collagenase type II for 30 min at 37℃.
We next investigated the effectiveness of the DP method for isolating active MSCs from muscle-derived primary cell populations. Using Method 3 (pronase and collagenase), we retrieved primary cell populations from skeletal muscle tissues of 3-week-old mice and separated the MSCs from the primary cell population using the DP method. The identity of the isolated MSCs was then confirmed by detecting the co-expression of Pax7 and MyoD. Most of the cells isolated from muscle-derived primary cell populations derived using the DP method exhibited double positivity for Pax7 and MyoD (Fig. 2A). The numbers of Pax7+/MyoD+ MSCs were significantly higher after DP (mean±SD, 78.19±9.72%) than before the cells were plated (mean±SD, 53.42±11.59%) (Fig. 2B). Based on these results, we confirmed that applying the DP method post-dissociation of skeletal muscle tissues enhances the selection of active MSCs from muscle-derived primary cell populations.
To investigate the effect of mouse age on the isolation of large numbers of active MSCs from skeletal muscle tissues, putative MSCs were collected from skeletal muscle tissues derived from 1-, 2-, 3-, and 4-week-old mice by applying DP method post-dissociation of skeletal muscle tissues through Method 3. For each sample, active MSCs were quantified by double-staining with Pax7 and MyoD. As shown in Fig. 3, in all replicates, a significantly larger number of Pax7+/MyoD+ MSCs could be harvested from 2-week-old mice (mean±SD, 7.84±2.84×105 cells) than from 1-week-old (mean±SD, 3.19±0.42×105 cells), 3-week-old (mean±SD, 1.03±0.14×105 cells), or 4-week-old mice (mean±SD, 1.27±0.41×105 cells). Moreover, the application of DP to 2-week-old mouse samples increased the percentage of Pax7+/MyoD+ MSCs to 88.38±3.18% (mean±SD). These results indicate that active MSCs can be isolated more effectively from skeletal muscle tissues derived from 2-week-old mice.
Here, we report a new protocol for high-yield isolation of active MSCs from skeletal muscle tissues in mice. A protocol generating muscle-derived primary cell population containing active MSCs at the highest proportion from skeletal muscle tissues and then an another protocol collecting precisely active MSCs from muscle-derived primary cell population was established in mice. In addition, sequential combination of these two protocols showed the best isolation yield of active MSCs with competence differentiating into the myotubes (Fig. S2) in the application to the skeletal tissues derived from 2-week-old mice. Accordingly, high-yield active MSCs isolated by the application of DP method post-dissociation of 2-week-old mouse-derived skeletal muscle tissues through digestion in 0.1% (w/v) pronase for 5 min at 37℃ after incubation in 0.2% (w/v) collagenase type II for 30 min at 37℃ will play a pivotal role in overcoming previous hurdles resulted from isolation of low-yield active MSCs from mouse skeletal muscle tissues, resulting in stimulating actively researches related with active MSCs.
To dissociate skeletal muscle tissues, we used different combinations of digestion enzymes. Collagenase was fundamentally used in all digestion methods to digest skeletal muscle tissues. In Method 1, we used collagenase only, in Method 2, collagenase and trypsin, and in Method 3, collagenase and pronase. The largest amount of active MSCs retrieved from primary cell populations was from skeletal muscle tissues that were digested by sequentially treating them with collagenase and pronase (Method 3) (Fig. 1). Thus, we speculate that pronase may play a crucial role in releasing active MSCs from histological networks in the skeletal muscle tissues, potentially by completely dismantling three-dimensional networks established from cell-to-cell interactions and extracellular matrix (ECM) macromolecules essential for skeletal muscle tissue formation. This speculation was based on a previous finding that the activities of collagenase and trypsin are restricted to cleave in the collagen network or the specific peptide sequence between serine and arginine (31, 32), whereas cleavage by pronase is random and does not depend on specific peptide sequences (32, 33).
As shown in Fig. 2, the purity of active MSCs was improved by applying the DP method to selectively enrich active MSCs from muscle-derived primary cell popula-tions. By taking advantage of the quick attachment of myocytes and myofibroblasts to the surface of culture plates, active MSCs could be easily harvested because they did not adhere as quickly as the other cells and remained floating in the medium. A previous study showed that MSCs localized in muscle tissues were quiescent and did not express any ECM proteins, whereas when separated from muscle tissues, the quiescent MSCs were immediately activated and over 42 hours gradually synthesized ECM proteins such as fibronectin (34). Moreover, the presence or absence of ECM proteins on the cell surface can influence the attachment of cells to the culture plate surface (35, 36). Therefore, the quantitative alteration of ECM protein expression on the surface of MSCs in the process of DP may contribute greatly to the effective selection of active MSCs from muscle-derived primary cell populations.
The highest yield of active MSCs was derived from the skeletal tissues of 2-week-old mice (Fig. 3). In general, hypertrophic growth in mice progresses from the time of birth up to postnatal week 6 through strong stimulation of myogenesis (37, 38). During this period, active MSCs differentiate from muscle progenitor cells and actively proliferate, resulting in the formation of many myonuclei, which increase myofiber size (37, 38). Previous studies have demonstrated the presence of an average of 50, 100, and 50 myonuclei in a myofiber of 1-, 2-, and 3-week-old mice, respectively, indicating that 2-week-old mice have the largest number of active MSCs in their muscles (39), which strongly supports our finding. Accordingly, we speculate that the largest number of active MSCs was isolated from the muscle tissues of 2-week-old mice because active MSCs are more abundant in muscle tissues in mice of that age than in the muscle tissues of 1- and 3-week-old mice.
In this study, we could develop the convenient and economic isolation protocol of active MSCs from skeletal muscle tissues by reducing the time of exposure to proteolytic enzymes, simplifying differential plating steps, and not using ECM proteins. In previously reported isolation protocols, skeletal muscle tissues are exposed to proteolytic enzymes for 90∼145 min to release mononuclear cells from skeletal muscle tissues (25, 40, 41). However, in this study, the exposure time of the proteolytic enzymes can be reduced to 35 min using collagenase type II and pronase. In addition, most of the reported isolation protocols of active MSCs have complicated differential plating consisting of three to eight steps (25, 40, 41). In our isolation protocol, active MSCs could be isolated from skeletal muscle tissue-derived primary cell populations through one differential plating step. Also, ECM proteins, such as collagen and Matrigel, are used to remove fibroblasts or induce adherence of active MSCs on ECM proteins in general (25, 40, 41). However, we could isolate a similar or higher yield of active MSCs without ECM proteins compared to the retrieval yield of active MSCs derived from most of the reported isolation protocols.
In conclusion, we established a high-yield isolation protocol for collecting active MSCs by applying the DP method to primary cell populations harvested from skeletal muscle tissues of 2-week-old mice. The skeletal muscle tissues were digested with collagenase type II for 30 min at 37℃ and then with pronase for 5 min at 37℃. This protocol is expected to play a pivotal role in overcoming previous hurdles often encountered when attempting to isolate active MSCs from mouse skeletal muscle tissues. By enabling the economically efficient, high-yield isolation of active MSCs from mouse skeletal muscle tissues, our protocol will contribute greatly to boosting muscle-related research.
Supplementary data including one table and two figures can be found with this article online at https://doi.org/10.15283/ijsc21179
Acknowledgments
This work was supported by the Technology Innovation Program (Alchemist Project, 20012384) funded by the Ministry of Trade, Industry & Energy (MOTIE, Korea), NRF (National Research Foundation of Korea) grant funded by the Korean Government (NRF-2016-Global Ph.D. Fellowship Program), and 2018 Research grant (PoINT) from Kangwon National University.
Notes
Potential Conflict of Interest
Author Na Rae Han is employed by SCBIO Co., Ltd and Jung Im Yun and Seung Tae Lee are employed by KustoGen Inc. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Chen W, Datzkiw D, Rudnicki MA. 2020; Satellite cells in ageing: use it or lose it. Open Biol. 10:200048. DOI: 10.1098/rsob.200048. PMID: 32428419. PMCID: PMC7276531. PMID: cfe5a68370ff46d6af0311c09747b508. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85084961672&origin=inward.
2. Han WM, Anderson SE, Mohiuddin M, Barros D, Nakhai SA, Shin E, Amaral IF, Pêgo AP, García AJ, Jang YC. 2018; Synthetic matrix enhances transplanted satellite cell engraftment in dystrophic and aged skeletal muscle with comorbid trauma. Sci Adv. 4:eaar4008. DOI: 10.1126/sciadv.aar4008. PMID: 30116776. PMCID: PMC6093653. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85052200928&origin=inward.
3. Cho CH, Lee KJ, Lee EH. 2018; With the greatest care, stromal interaction molecule (STIM) proteins verify what skeletal muscle is doing. BMB Rep. 51:378–387. DOI: 10.5483/BMBRep.2018.51.8.128. PMID: 29898810. PMCID: PMC6130827. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85055079049&origin=inward.
4. Peake JM, Neubauer O, Della Gatta PA, Nosaka K. 2017; Muscle damage and inflammation during recovery from exercise. J Appl Physiol (1985). 122:559–570. DOI: 10.1152/japplphysiol.00971.2016. PMID: 28035017. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85014792249&origin=inward.
5. Delos D, Maak TG, Rodeo SA. 2013; Muscle injuries in athletes: enhancing recovery through scientific understanding and novel therapies. Sports Health. 5:346–352. DOI: 10.1177/1941738113480934. PMID: 24459552. PMCID: PMC3899907. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84879182162&origin=inward.
6. Thooyamani AS, Mukhopadhyay A. 2021; PDGFRα mediated survival of myofibroblasts inhibit satellite cell proliferation during aberrant regeneration of lacerated skeletal muscle. Sci Rep. 11:63. DOI: 10.1038/s41598-020-79771-4. PMID: 33420132. PMCID: PMC7794387. PMID: eb684f04a9c94611bd1374ec7155475d. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85098994697&origin=inward.
7. Ribeiro AF Jr, Souza LS, Almeida CF, Ishiba R, Fernandes SA, Guerrieri DA, Santos ALF, Onofre-Oliveira PCG, Vainzof M. 2019; Muscle satellite cells and impaired late stage regeneration in different murine models for muscular dystrophies. Sci Rep. 9:11842. DOI: 10.1038/s41598-019-48156-7. PMID: 31413358. PMCID: PMC6694188. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85070946308&origin=inward.
8. Verma M, Asakura Y, Murakonda BSR, Pengo T, Latroche C, Chazaud B, McLoon LK, Asakura A. 2018; Muscle satellite cell cross-talk with a vascular niche maintains quiescence via VEGF and Notch signaling. Cell Stem Cell. 23:530–543.e9. DOI: 10.1016/j.stem.2018.09.007. PMID: 30290177. PMCID: PMC6178221. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85053859628&origin=inward.
9. Jeong JY, Kim JM, Rajesh RV, Suresh S, Jang GW, Lee KT, Kim TH, Park M, Jeong HJ, Kim KW, Cho YM, Lee HJ. 2013; Comparison of gene expression levels of porcine satellite cells from postnatal muscle tissue during differenti-ation. Reprod Dev Biol. 37:219–224. DOI: 10.12749/RDB.2013.37.4.219.
10. Baghdadi MB, Tajbakhsh S. 2018; Regulation and phylogeny of skeletal muscle regeneration. Dev Biol. 433:200–209. DOI: 10.1016/j.ydbio.2017.07.026. PMID: 28811217. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85029700470&origin=inward.
11. Forcina L, Cosentino M, Musarò A. 2020; Mechanisms regulating muscle regeneration: insights into the interrelated and time-dependent phases of tissue healing. Cells. 9:1297. DOI: 10.3390/cells9051297. PMID: 32456017. PMCID: PMC7290814. PMID: 7bfbfd899636487584d16c4b6e2f627c. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85085538839&origin=inward.
12. Larrick JW, Larrick JW, Mendelsohn AR. 2016; Reversal of aged muscle stem cell dysfunction. Rejuvenation Res. 19:423–429. DOI: 10.1089/rej.2016.1875. PMID: 27612523. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84992200362&origin=inward.
13. Pini V, Morgan JE, Muntoni F, O'Neill HC. 2017; Genome editing and muscle stem cells as a therapeutic tool for muscular dystrophies. Curr Stem Cell Rep. 3:137–148. DOI: 10.1007/s40778-017-0076-6. PMID: 28616376. PMCID: PMC5445179. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85045943044&origin=inward.
14. Zhu P, Wu F, Mosenson J, Zhang H, He TC, Wu WS. 2017; CRISPR/Cas9-mediated genome editing corrects dystrophin mutation in skeletal muscle stem cells in a mouse model of muscle dystrophy. Mol Ther Nucleic Acids. 7:31–41. DOI: 10.1016/j.omtn.2017.02.007. PMID: 28624206. PMCID: PMC5363682. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85020720585&origin=inward.
15. Alway SE, Myers MJ, Mohamed JS. 2014; Regulation of satellite cell function in sarcopenia. Front Aging Neurosci. 6:246. DOI: 10.3389/fnagi.2014.00246. PMID: 25295003. PMCID: PMC4170136. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84907942844&origin=inward.
16. Said RS, Mustafa AG, Asfour HA, Shaqoura EI. 2017; Myogenic satellite cells: biological milieu and possible clinical applications. Pak J Biol Sci. 20:1–11. DOI: 10.3923/pjbs.2017.1.11. PMID: 29023009. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85006356043&origin=inward.
17. Post MJ, Levenberg S, Kaplan DL, Genovese N, Fu J, Bryant CJ, Negowetti N, Verzijden K, Moutsatsou P. 2020; Scientific, sustainability and regulatory challenges of cultured meat. Nat Food. 1:403–415. DOI: 10.1038/s43016-020-0112-z. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85088867952&origin=inward.
18. Choi KH, Yoon JW, Kim M, Lee HJ, Jeong J, Ryu M, Jo C, Lee CK. 2021; Muscle stem cell isolation and in vitro culture for meat production: a methodological review. Compr Rev Food Sci Food Saf. 20:429–457. DOI: 10.1111/1541-4337.12661. PMID: 33443788. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85096673947&origin=inward.
19. Stephens N, Di Silvio L, Dunsford I, Ellis M, Glencross A, Sexton A. 2018; Bringing cultured meat to market: technical, socio-political, and regulatory challenges in cellular agriculture. Trends Food Sci Technol. 78:155–166. DOI: 10.1016/j.tifs.2018.04.010. PMID: 30100674. PMCID: PMC6078906. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85048566593&origin=inward.
20. Treich N. 2021; Cultured meat: promises and challenges. Environ Resour Econ (Dordr). doi: 10.1007/s10640-021-00551-3. [Epub ahead of print]. DOI: 10.1007/s10640-021-00551-3. PMID: 33758465. PMCID: PMC7977488. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85103064615&origin=inward.
21. Srutee R, R S S, Uday S A. 2021; Clean meat: techniques for meat production and its upcoming challenges. Anim Biotechnol. doi: 10.1080/10495398.2021.1911810. [Epub ahead of print]. DOI: 10.1080/10495398.2021.1911810. PMID: 33947302. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85105328829&origin=inward.
22. Yin H, Price F, Rudnicki MA. 2013; Satellite cells and the muscle stem cell niche. Physiol Rev. 93:23–67. DOI: 10.1152/physrev.00043.2011. PMID: 23303905. PMCID: PMC4073943. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84872281980&origin=inward.
23. McKay BR, Nederveen JP, Fortino SA, Snijders T, Joanisse S, Kumbhare DA, Parise G. 2020; Brain-derived neurotrophic factor is associated with human muscle satellite cell differentiation in response to muscle-damaging exercise. Appl Physiol Nutr Metab. 45:581–590. DOI: 10.1139/apnm-2019-0501. PMID: 31661631. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85085904103&origin=inward.
24. Shahini A, Vydiam K, Choudhury D, Rajabian N, Nguyen T, Lei P, Andreadis ST. 2018; Efficient and high yield isolation of myoblasts from skeletal muscle. Stem Cell Res. 30:122–129. DOI: 10.1016/j.scr.2018.05.017. PMID: 29879622. PMCID: PMC6090567. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85047855425&origin=inward.
25. Yoshioka K, Kitajima Y, Okazaki N, Chiba K, Yonekura A, Ono Y. 2020; A modified pre-plating method for high-yield and high-purity muscle stem cell isolation from human/mouse skeletal muscle tissues. Front Cell Dev Biol. 8:793. DOI: 10.3389/fcell.2020.00793. PMID: 32903486. PMCID: PMC7438441. PMID: 76aca5e166e449acb21aac192da73336. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85089938198&origin=inward.
26. Motohashi N, Asakura Y, Asakura A. 2014; Isolation, culture, and transplantation of muscle satellite cells. J Vis Exp. (86):50846. DOI: 10.3791/50846. PMID: 24747722. PMCID: PMC4131689. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84898746462&origin=inward.
27. Benedetti A, Cera G, De Meo D, Villani C, Bouche M, Lozanoska-Ochser B. 2021; A novel approach for the isolation and long-term expansion of pure satellite cells based on ice-cold treatment. Skelet Muscle. 11:7. DOI: 10.1186/s13395-021-00261-w. PMID: 33731194. PMCID: PMC7968259. PMID: 3a65cd8f1149438a87f3bea3700016a6. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85102718477&origin=inward.
28. Gromova A, Tierney MT, Sacco A. 2015; FACS-based satellite cell isolation from mouse hind limb muscles. Bio Protoc. 5:e1558. DOI: 10.21769/BioProtoc.1558. PMID: 27668269. PMCID: PMC5034768.
29. Maesner CC, Almada AE, Wagers AJ. 2016; Established cell surface markers efficiently isolate highly overlapping populations of skeletal muscle satellite cells by fluorescence-activated cell sorting. Skelet Muscle. 6:35. DOI: 10.1186/s13395-016-0106-6. PMID: 27826411. PMCID: PMC5100091. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84999158343&origin=inward.
30. Syverud BC, Lee JD, VanDusen KW, Larkin LM. 2014; Isolation and purification of satellite cells for skeletal muscle tissue engineering. J Regen Med. 3:117. DOI: 10.4172/2325-9620.1000117. PMID: 26413555. PMCID: PMC4582791.
31. Singh A, Benjakul S. 2018; Proteolysis and its control using protease inhibitors in fish and fish products: a review. Compr Rev Food Sci Food Saf. 17:496–509. DOI: 10.1111/1541-4337.12337. PMID: 33350077. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85042024856&origin=inward.
32. Miersch C, Stange K, Röntgen M. 2018; Effects of trypsinization and of a combined trypsin, collagenase, and DNase digestion on liberation and in vitro function of satellite cells isolated from juvenile porcine muscles. In Vitro Cell Dev Biol Anim. 54:406–412. DOI: 10.1007/s11626-018-0263-5. PMID: 29785535. PMCID: PMC5997727. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85047194017&origin=inward.
33. Brown NK, Meade JR, Wang J, Marino SR. 2017; Reanalysis of the role of pronase treatment of B cells in the flow cytometric crossmatch assay: Fc receptor is not the primary target. Hum Immunol. 78:704–709. DOI: 10.1016/j.humimm.2017.10.002. PMID: 28987959. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85030788393&origin=inward.
34. Bentzinger CF, Wang YX, von Maltzahn J, Soleimani VD, Yin H, Rudnicki MA. 2013; Fibronectin regulates Wnt7a signaling and satellite cell expansion. Cell Stem Cell. 12:75–87. DOI: 10.1016/j.stem.2012.09.015. PMID: 23290138. PMCID: PMC3539137. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84872009392&origin=inward.
35. Park MH, Kim MS, Yun JI, Choi JH, Lee E, Lee ST. 2018; Integrin heterodimers expressed on the surface of porcine spermatogonial stem cells. DNA Cell Biol. 37:253–263. DOI: 10.1089/dna.2017.4035. PMID: 29369695. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85045425624&origin=inward.
36. Kim SJ, Kim MS, Park HJ, Lee H, Yun JI, Lim HW, Lee ST. 2020; Screening of integrins localized on the surface of human epidermal melanocytes. In Vitro Cell Dev Biol Anim. 56:435–443. DOI: 10.1007/s11626-020-00471-4. PMID: 32572848. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85086778940&origin=inward.
37. Relaix F, Zammit PS. 2012; Satellite cells are essential for skeletal muscle regeneration: the cell on the edge returns centre stage. Development. 139:2845–2856. DOI: 10.1242/dev.069088. PMID: 22833472. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84864317860&origin=inward.
38. Butchart LC, Fox A, Shavlakadze T, Grounds MD. 2016; The long and short of non-coding RNAs during post-natal growth and differentiation of skeletal muscles: focus on lncRNA and miRNAs. Differentiation. 92:237–248. DOI: 10.1016/j.diff.2016.05.003. PMID: 27292314. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84998694304&origin=inward.
39. White RB, Biérinx AS, Gnocchi VF, Zammit PS. 2010; Dynamics of muscle fibre growth during postnatal mouse develop-ment. BMC Dev Biol. 10:21. DOI: 10.1186/1471-213X-10-21. PMID: 20175910. PMCID: PMC2836990. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=77649309210&origin=inward.
40. Gharaibeh B, Lu A, Tebbets J, Zheng B, Feduska J, Crisan M, Péault B, Cummins J, Huard J. 2008; Isolation of a slowly adhering cell fraction containing stem cells from murine skeletal muscle by the preplate technique. Nat Protoc. 3:1501–1509. DOI: 10.1038/nprot.2008.142. PMID: 18772878. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=51649123800&origin=inward.
41. Lavasani M, Lu A, Thompson SD, Robbins PD, Huard J, Niedernhofer LJ. 2013; Isolation of muscle-derived stem/progenitor cells based on adhesion characteristics to collagen-coated surfaces. Methods Mol Biol. 976:53–65. DOI: 10.1007/978-1-62703-317-6_5. PMID: 23400434. PMCID: PMC3731996. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84880576445&origin=inward.
Fig. 1
Percentage of active MSCs in muscle-derived primary cell populations isolated using different muscle dissociation protocols. Each primary cell population was derived from skeletal muscle tissues of 3-week-old mice according to the experimental design described in Fig. S1. Muscle-derived primary cell populations were double-stained with antibodies detecting Pax7 (red) and MyoD (green), which are expressed simultaneously in active MSCs. The percentage of double-stained cells was determined by multiplying the number of double-stained cells divided by the total number of cells with 100. MSCs that exhibit positive double-staining for Pax7 and MyoD were observed within muscle-derived primary cell populations, irrespective of the muscle dissociation protocol (A). In all experimental replicates, primary cell populations obtained using Method 3 exhibited the highest percentage of active MSCs (B). Representative images of the co-expression of MyoD and Pax7 in active MSCs are displayed. Nuclear counterstaining was performed using 4’,6-diamidino-2-phenylindole (DAPI; blue), n=5. Scale bars represent 50 μm. All data are presented as the mean (dotted line) of five independent experiments.
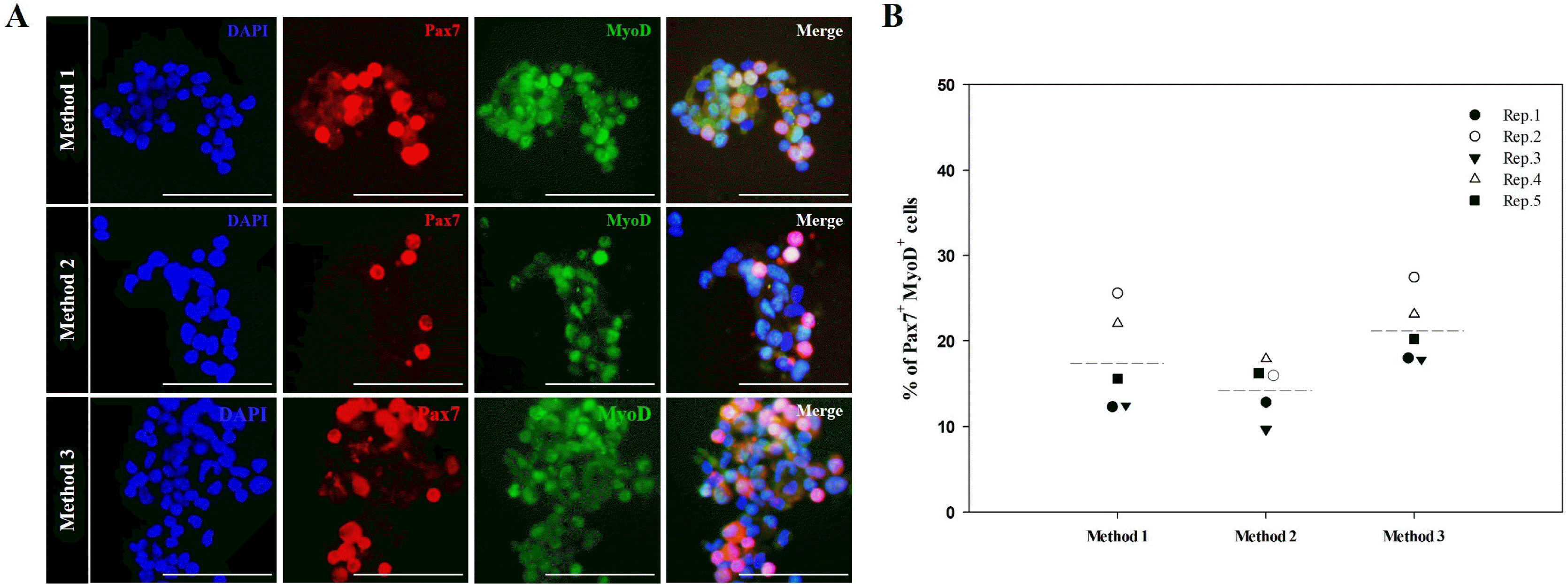
Fig. 2
Effects of DP method on improving active MSC purity from muscle-derived primary cell populations. The DP method was used to retrieve putative MSCs from muscle-derived primary cell populations of 3-week-old mice generated using Method 3. The cells were double-stained with antibodies against Pax7 (red) and MyoD (green), which are expressed simultaneously in active MSCs, and nuclear counterstaining was performed using DAPI (blue). Co-localization of Pax7 and MyoD in the nuclear region indicates active MSCs (A). The percentage of double-stained cells (indicating active MSCs) was determined by multiplying the number of double-stained cells divided by the total number of cells with 100. The percentage of active MSCs co-expressing Pax7 and MyoD was significantly higher in populations subjected to the DP method (B). All figures shown in (A) are representative images of the co-expression of Pax7 and MyoD in active MSCs, and all data shown in (B) represent the mean±SD of three independent experiments. *p<0.05, n=3 in (A). Scale bars represent 50 μm.
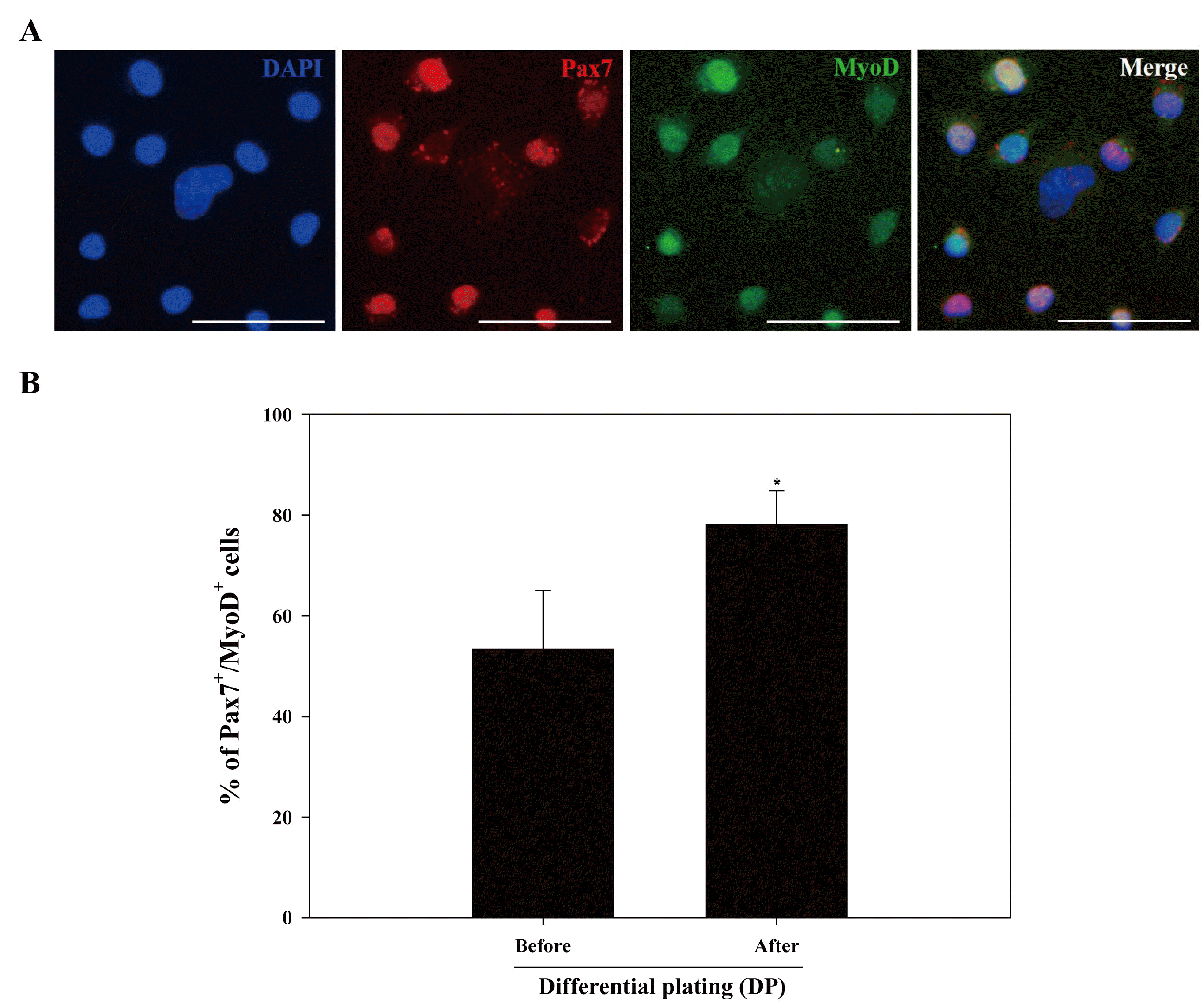
Fig. 3
Effects of difference in mouse age on isolating active MSCs from skeletal muscle tissues. Muscle-derived primary cell populations were retrieved from skeletal muscle tissues from 1-, 2-, 3-, and 4-week-old mice using Method 3. Collection of putative MSCs from each muscle-derived primary cell population was conducted using the DP method. All putative MSCs were double-stained with anti-Pax7 and anti-MyoD antibodies. Total cell numbers and the numbers of double-stained cells were counted. In all repeated experiments, the highest number of cells double-stained for Pax7 and MyoD was consistently observed in putative MSCs derived from 2-week-old mice compared to those from 1-, 3-, and 4-week-old mice. All data are presented as the mean (dotted line) of three independent experiments. *p<0.05.
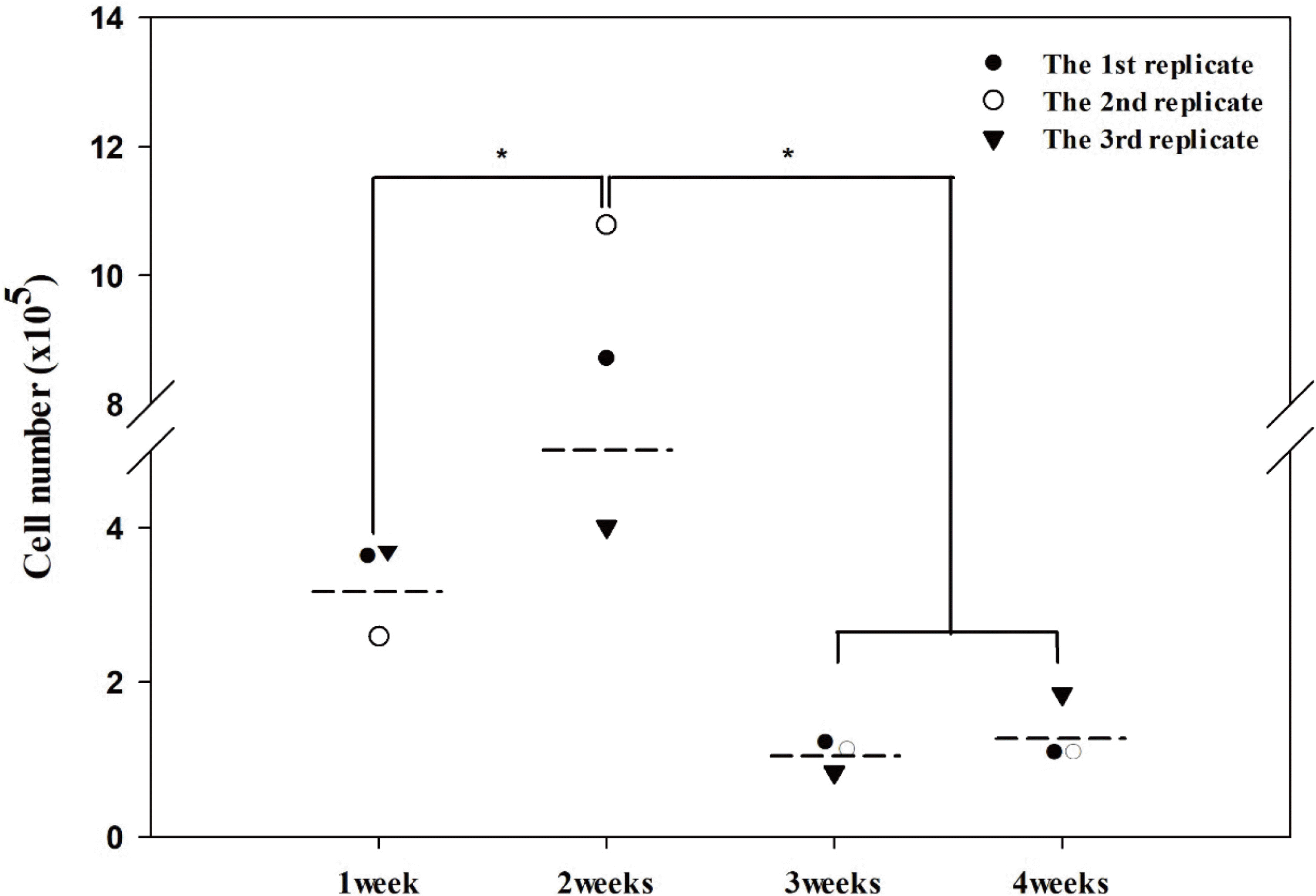
Table 1
Primary and secondary antibody list




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download