Abstract
Objective
To evaluate differences in the adhesion levels of the most common oral pathogens, Streptococcus mutans and Porphyromonas gingivalis, in human saliva-derived microcosm biofilms with respect to time and raw materials of orthodontic brackets.
Methods
The samples were classified into three groups of bracket materials 1) monocrystalline alumina ceramic (CR), 2) stainless steel metal (SS), and 3) polycarbonate plastic (PL), and a hydroxyapatite (HA) group was used to mimic the enamel surface. Saliva was collected from a healthy donor, and saliva-derived biofilms were grown on each sample. A real-time polymerase chain reaction was performed to quantitatively evaluate differences in the attachment levels of total bacteria, S. mutans and P. gingivalis at days 1 and 4.
Results
Adhesion of S. mutans and P. gingivalis to CR and HA was higher than the other bracket materials (SS = PL < CR = HA). Total bacteria demonstrated higher adhesion to HA than to bracket materials, but no significant differences in adhesion were observed among the bracket materials (CR = SS = PL < HA). From days 1 to 4, the adhesion of P. gingivalis decreased, while that of S. mutans and total bacteria increased, regardless of material type.
Orthodontic treatments using various fixed appliances are commonly used to improve function and facial esthetics in children and adults. Enamel decalcification and gingival inflammation are the common complications associated with orthodontic treatments using fixed appliances.1 These side effects are primarily a result of increased biofilm formation and microbial changes of oral biofilms.1 In particular, Streptococcus mutans is the primary causative bacteria of enamel decalcification due to its ability to promote biofilm development, produce lactic acid, and resist against acidic environment.2 Porphyromonas gingivalis is putatively involved in gingival and/or periodontal inflammation due to its ability to disturb the immune system of periodontal tissues.2
Among the various fixed appliances, orthodontic brackets are associated with an increased risk of orthodontic complications because they promote biofilm development and changes in biofilm composition by hampering the maintenance of oral hygiene and increasing the retention sites for oral bacteria.2,3 In addition, brackets and their peripheral area constitute sites for oral biofilm development because of the irregular surface and increased wettability associated with the bracket materials used.4,5 Their uneven surface protects bacteria against hydro-dynamic shear forces, and increased wettability induces strong adhesion of oral bacteria to underlying surfaces, leading to rapid biofilm development at these locations.4,5
Previous studies have reported biofilm development on various commercial orthodontic brackets using a single bacterial species model;5,6 however, these studies could not provide accurate information on how brackets influence biofilm development as commercial brackets of different sizes and designs were used in different studies. Additionally, a biofilm model with a single bacterial species cannot correctly simulate the human oral cavity's complex and diverse microbial environment.7 Considering that human saliva contains various bacteria from different microbial communities that adhere to the intraoral surfaces, it is a useful source of bacterial inoculum that can be used to reproduce the oral biofilm responsible for highly prevalent orthodontic complications.8 However, till date, few studies have evaluated the interaction of bracket materials with the oral microflora using an actual human oral ecosystem. The objective of this in vitro study was to assess the compositional difference in S. mutans and P. gingivalis in saliva-derived biofilms using uniformly shaped specimens of raw orthodontic bracket materials.
Three bracket materials and hydroxyapatite were used: (1) ceramic [CR] (monocrystalline alumina, Al2O3; HUBIT Co., Ltd., Seoul, Korea), (2) stainless steel metal [SS] (SUS304; HUBIT Co., Ltd.), (3) plastic [PL] (polycarbonate; HUBIT Co., Ltd), and (4) hydroxyapatite [HA] (RD128-HA; BioSurface Tech Co., Bozeman, MT, USA). HA was used as the control group to simulate the enamel surface. Each material was prepared as a uniform disc-shaped specimen (thickness of 3.0 mm and diameter of 12.7 mm) that was used to grow microbial biofilms in environments similar to that of the oral cavity using a CDC biofilm reactor system (BioSurface Tech Co).9 Twenty-five specimens for each group were provided by the manufacturer. A total of 100 specimens were used. The surface morphology of the material was analyzed from one specimen randomly selected from each group. Surface roughness, surface wettability (water contact angle), and microbial tests were analyzed from the other specimens.
Surface roughness was evaluated by calculating the mean surface roughness within the specific area (450 × 450 × 50 μm). Using a confocal laser scanning microscope, each specimen was randomly measured at three points (LSM 5 Pascal; Carl Zeiss MicroImaging GmbH, Göttingen, Germany). Surface wettability was evaluated by calculating the specimens’ water contact angles using a video-based optic system (Phoenix 300; Surface Electro Optics, Suwon, Korea) as previously described.10 The surface morphology was analyzed at 3,000× magnification using a scanning electron microscope (SEM) (S-4700 microscope; Hitachi, Tokyo, Japan).
As an inoculum for multispecies biofilm development, we used saliva collected from a single donor because the microflora of a single donor is relatively constant, and saliva samples from multiple donors exhibit a large degree of microflora variation.11,12 A spitting method was used to collect unstimulated whole saliva from a healthy donor without active periodontal or caries lesions who had not taken antibiotics within the last three months, as previously described.5 The donor was not allowed to consume any food or drink or brush the teeth 12 hours before saliva collection. The collected saliva was stored at −80°C after being diluted to a concentration of 70% using sterile glycerol. The research protocol was approved by institutional review board of Seoul National University School of Dentistry (S-D20170021).
A basal mucin medium (BMM) containing porcine potassium chloride (2.5 g/L), gastric mucin (2.5 g/L), proteose peptone (2 g/L), yeast extract (1 g/L), trypticase peptone (1 g/L), cysteine hydrochloride (0.1 g/L), hemin (0.001 g/L), urea (10 mM), and glucose (10 mM) was used for making biofilm media as previously described.13 The saliva-derived biofilm was cultivated in a CDC biofilm reactor that mimicked the dynamic conditions of the oral cavity by providing the continuous and constant flow of fresh BMM at a defined rate. Six disc-shaped specimens from each of the four material groups were randomly mounted onto eight independent rods, each of which held three specimens. After sterilization of the rods with the mounted disc-shaped specimens, the CDC biofilm reactor was rotated at 60 rpm on a 37°C hot stir plate.14 After thawing, 5 mL of the stored saliva was added into the biofilm reactor as a biofilm inoculum, after which BMM continuously flowed to the reactor at 100 mL/h.15
To evaluate the differences in bacterial composition among the four material groups, 12 specimens (three sets of four specimens from the different materials groups) were collected from the reactor at days 1 (early biofilms) and 4 (mature biofilms), respectively, as mature biofilms can be obtained after 72 hours of incubation in the reactor.16 Each specimen was then moved into a conical tube and washed twice with phosphate-buffered saline (PBS, pH = 7.4). Then, the biofilm was dismounted from the specimen using sonication as previously described.15,17 The resulting cell suspension was spun down at 13,000 rpm for 10 minutes after washing with PBS. The CellEase Bacteria II Genomic DNA Extraction Kit (Biocosm, Osaka, Japan) was used to extract bacterial chromosomal DNA according to the manufacturer’s instructions.
Real-time polymerase chain reaction (PCR) (the iQ5 system; Bio-Rad, Hercules, CA, USA) was performed to quantify the target bacteria. The PCR mixtures consisted of 2 µL purified DNA from the specimens, 10 µL 2x iQ SYBR Green Supermix (Bio-Rad), and 100 pM primer. Distilled water was added to each PCR mixture to a final volume of 20 µL. Table 1 shows known specific PCR primers (provided from Bioneer, Daejeon, Korea) and the thermal cycling conditions for quantifying total bacteria, S. mutans and P. gingivalis.
A DNA standard curve was used to estimate the bacterial number in the biofilms as previously described.18 The DNA standard curve was constructed using purified PCR products from P. gingivalis KCOM 2797 and S. mutans ATCC 700610. All the microbiological experiments were performed in triplicate and independently repeated four times.
Two-way ANOVA (IBM® SPSS® Statistics, version 21; IBM Corp., Armonk, NY, USA) was performed to analyze the differences in the bacterial composition according to incubation time and material type. The Kruskal–Wallis and Mann–Whitney tests with Bonferroni correction (IBM® SPSS® Statistics, version 21) were used to evaluate the differences in surface roughness and water contact angle among the four materials. The significance level was set at α = 0.05.
Our results showed significant differences in surface roughness among the four materials (CR = SS < PL < HA, Table 2). A significant difference in water contact angle was also found; however, the water contact angle of CR was similar to that of HA. PL had the highest water contact angle followed by SS, CR, and HA (CR = HA < SS < PL, Table 2).
The results of surface morphology were not consistent with those of surface roughness. HA (Figure 1D) showed more micro-porosities, irregularities, and flaws on its surface compared to bracket materials. CR (Figure 1A) exhibited more irregular textures with narrower grooves and deeper pits among the three bracket materials, while PL (Figure 1C) showed a relatively even surface with broader and shallower grooves. SS (Figure 1B) had the smoothest texture.
Table 3 demonstrates the differences in the bacterial composition according to incubation time and material type. Our results showed that biofilm composition varied significantly according to incubation time and material type without interaction effects between them. Moreover, significant difference in the adhesion level of oral pathogens were shown according to the material type. CR and HA exhibited higher adhesion of oral pathogens than SS and PL, with S. mutans showing higher adhesion to CR and HA than to SS and PL (SS = PL < CR = HA, p < 0.05), whereas P. gingivalis showed higher adhesion to CR and HA than to SS (SS < CR = HA, p < 0.05). In addition, total bacteria showed higher adhesion to HA than bracket materials, but no significant difference was observed in total bacterial adhesion among the three bracket materials (CR = SS = PL < HA, p < 0.05, Table 3). Bacterial adhesion level significantly differed according to time.
Bracket placement leads to ecological changes in the microbial consortium in oral biofilms by shifting the bacterial amount, composition, and pathogenicity, leading to a higher incidence of enamel decalcification and gingivitis.2,3 Although studies have examined the interactions of oral bacteria with orthodontic brackets, the specific bracket type that provides a favorable environment for biofilm development remains unclear.4,19,20 A previous study using prefabricated brackets reported lower adhesion of S. mutans to ceramic brackets than to metal brackets, whereas another study showed the opposite results.20,21 This is because the differences in the size, material type, design, and relevant physicochemical surface properties of the brackets studied significantly influence bacterial adhesion and biofilm development.22 To overcome these discrepancies, other studies used raw materials, whereas most used a single bacterial species in a static biofilm model.4,5 In fact, in situ or in vivo studies are the best methods to investigate biofilm development on bracket materials, as the oral cavity is a highly heterogeneous and dynamic system containing diverse microorganisms.7 However, the use of an oral microbial consortium from human volunteers leads to practical problems, such as ethical limitations, compliance, and diet control.11 Many in vitro biofilm models have been proposed to simulate and reproduce the complicated in vivo environment. In vitro studies generally use three types of oral biofilm models: pure culture (single species), defined consortium (simple or multi-species), and microcosm (human saliva or dental plaque).23 However, most studies have used a pure culture or defined consortium biofilm model to adequately control various variables and be readily reproducible and identifiable, although these models could not completely mimic the conditions of the actual oral environment.5,6,11 Recently, a saliva-derived microcosm model has been introduced for the in vitro cultivation of oral biofilms to preserve most of the heterogeneity and complexity of oral biofilms in vivo.23 Thus, the present study assessed differences in the adhesion of oral pathogens to bracket materials using a microcosm biofilm model derived from human saliva.
Surface properties such as surface roughness, wettability, and morphology significantly contribute to bacterial adhesion and biofilm development.24,25 Rough surfaces increase the adhesion area for biofilm development and protect biofilms from external shear forces.26 In our study, surface roughness was evaluated using a confocal laser scanning microscopy. Surface wettability is another critical factor for bacterial adhesion to underlying materials, as a material with higher surface wettability attracts more bacteria to its surface than a material with lower surface wettability due to its higher nonspecific physicochemical interactions (acid-base, van der Waals, and electrostatic interactions) between the surface and bacteria.26 Surface wettability is measured using the contact angle of a probe liquid on the surface, where a small contact angle corresponds to a higher surface wettability, and a larger contact angle corresponds to a lower surface wettability.27 Surface morphology also significantly influences bacterial adhesion because early bacterial colonization on enamel surfaces starts at surface irregularities (grooves, pits, and defects). The bacteria attached to irregular surfaces are substantially protected from external shear forces.24 In the present study, surface morphology was qualitatively examined using SEM.
During biofilm development in the oral cavity, the adhesion of early colonizers is the first step in the pathogenesis of oral infectious diseases. It occurs through non-specific and specific microbial-substrate adhesions.7 During this stage, surface properties contribute to the adhesion of early colonizers because they directly interact with the underlying surfaces.25 In particular, surface roughness is reported to play a substantial role in the adhesion of early colonizers, such as S. mutans.5,28
In this study, S. mutans adhesion was not significantly influenced by surface roughness. Although, the surface roughness of CR was lower than that of HA, no significant differences in S. mutans adhesion between HA and CR. In addition, S. mutans showed higher adhesion to CR than SS or PL, despite CR’s similar surface roughness to that of SS and its lower surface roughness than that of PL. This is because surface roughness above a specific limit cannot significantly influence bacterial adhesion in multi-species biofilms. In previous studies, S. mutans adhesion sharply increased until the surface roughness threshold of 0.2 μm was reached, and there were no significant differences in the adhesion of S. mutans when the surface roughness was above 0.35 μm.17,29 In our study, the average surface roughness of all the materials was more than 0.55 μm (Table 2); therefore, the effects of surface roughness on S. mutans adhesion might be minimal.
Furthermore, the porcine gastric mucin we used in the BMM coated the underlying surface of the specimen in a way similar to how salivary pellicles coat rough surfaces. This could be one of the reasons for the insignificant relation between surface roughness and bacterial adhesion. Mucin coating may conceal the surface properties of biomaterials and their effects on bacterial adhesion, especially surface roughness. A previous study showed that adsorbed mucin competes with S. mutans for the HA-binding sites and inhibits its adhesion to HA.30 Thus, surface wettability and morphology might have been the two significant factors influencing the adhesion of S. mutans in the present study.
S. mutans adhesion to HA and CR was higher than that to SS and PL (Table 3), with this likely explained by the lower water contact angle of HA and CR relative to that of SS and PL, considering the inverse relationship between surface wettability and water contact angle.4 This is because surface wettability affects S. mutans adhesion to underlying surfaces through the physicochemical interactions between the material surface and bacteria.26 In addition, surface morphology may also partly lead to higher S. mutans adhesion to HA (Figure 1D) and CR (Figure 1A) than to SS (Figure 1B) and PL (Figure 1C). The surface morphology (e.g., the number of grooves, pits, and irregularities) affects the adhesion of early colonizers, such as S. mutans.24 Because surface roughness is expressed as the mean of the depth of the randomly measured surfaces,29 it cannot fully explain differences in the number of grooves and pits. The higher S. mutans adhesion to HA and CR than to SS and PL may be because HA and CR had more irregularities (pores and flaws) on their surfaces than SS and PL (Figure 1).
With smaller surface wettability (higher water contact angle) than that of SS, PL exhibited an S. mutans adhesion similar to SS. The surface irregularities of PL (Figure 1C) were more than those of SS (Figure 1B). This could compensate for the smaller surface wettability and result in no significant difference in S. mutans adhesion between SS and PL.
Similar to S. mutans adhesion, P. gingivalis adhesion to HA and CR was higher than SS (Table 3). This may be explained by the adhesions between the early (S. mutans) and late colonizers (P. gingivalis). When biofilm formation begins, the early colonizers attach first, followed by the adhesion of the middle and late colonizers.28,31 In particular, the successful colonization of the late colonizers mainly depends on their adhesion to the already adhered early colonizers.28,31 Therefore, the adhesion of the late colonizers may be more affected by the adhesion of the early colonizers than by the surface properties of the materials.
This study showed that total bacteria adhered more to HA than to the three bracket materials, but their adhesion did not significantly differ among the bracket materials (Table 3). This is because other factors and the surface properties of the underlying materials may significantly affect the adhesion of total bacteria. Early biofilm develops after the deposition of a salivary pellicle and cell aggregation on the surface, and biofilm maturation and bacterial growth proceed via an intra- and inter-generic co-aggregation of planktonic bacteria to the already formed biofilm. Although surface properties may partly contribute to the adhesion of total bacteria, intercellular interactions during biofilm maturation may mask the influence of surface properties on the adhesion of total bacteria to bracket materials. Our study showed a significant difference in the biofilm composition of the oral pathogens without a significant difference in the adhesion of total bacteria among the three bracket materials (Table 3).
Additionally, this study demonstrated that the incubation time significantly affected biofilm composition, regardless of the material type. S. mutans and total bacteria adhesion increased with time (day 1 < day 4), whereas P. gingivalis adhesion decreased (day 1 > day 4, Table 3). The different adhesion patterns between S. mutans and P. gingivalis with time may be associated with their growth characteristics in the presence of oxygen. Streptococci are dominant bacteria in oral biofilms, and most streptococci are facultative anaerobes.32 Because the CDC biofilm reactor used in this study was designed to supply continuous oxygen to mimic the aerobic conditions of the supra-gingival status, the growth and colonization of total bacteria and S. mutans were probably not significantly influenced by the presence of oxygen. Therefore, S. mutans and total bacteria adhesion may increase with time. In contrast, the growth of P. gingivalis in the biofilm reactor was probably significantly inhibited over time because of its obligate anaerobic nature. This is also consistent with the results of a previous multi-species biofilm study using orthodontic adhesives.15
Taken together, our study demonstrated that the composition of oral pathogens in bracket biofilms was significantly influenced by material type. Considering that the increased ratio of pathogenic bacteria (S. mutans or P. gingivalis) in the biofilm due to a change in a specific oral environment is more closely associated with dental caries or periodontal disease, respectively, than the amount of total bacteria,33,34 our results suggest that the potential risk of dental caries and gingival inflammation may be clinically different depending on the bracket material type, despite a similar amount of total biofilms. In particular, higher adhesion of the known oral pathogens to CR than SS and PL suggests that CR brackets may not be advantageous in the management of common orthodontic complications, specifically in patients with poor oral hygiene.
Despite the findings of our results, the present study showed limitations. First, the saliva sample was obtained from a subject with good oral health. Therefore, this sample may not accurately represent the oral microbiome of various orthodontic patients. However, saliva from a single donor produces consistent results because the microflora of a single donor is constant.11,12 Additional in vitro and in vivo studies according to various environmental conditions (age, sex, and oral hygiene status) may be needed to verify the relationships between orthodontic materials and orthodontic complications. Second, an in vitro saliva-derived biofilm model may reproduce the oral environment but may not accurately simulate the diverse and complex environment of the human oral cavity. Thus, a further investigation with an in vivo or in situ biofilm model using a split-mouth design is warranted to overcome the limitations of the in vitro study.
This study showed significant differences in the composition of pathogenic bacteria among biofilms formed on bracket materials. The use of CR brackets may be related to orthodontic side effects, such as enamel decalcification and gingivitis, because of the higher adhesion of S. mutans and P. gingivalis to CR than to other bracket materials. Therefore, orthodontists should carefully monitor oral hygiene status, specifically when CR brackets are used in orthodontic patients with poor oral hygiene.
ACKNOWLEDGEMENTS
Porphyromonas gingivalis KCOM 2797 was kindly obtained from Korean Collection for Oral Microbiology (Gwangju, Korea).
This work was supported by the Technology Innovation Program (20001155) funded by the Ministry of Trade, Industry & Energy (MOTIE, Korea).
REFERENCES
1. Low B, Lee W, Seneviratne CJ, Samaranayake LP, Hägg U. 2011; Ultrastructure and morphology of biofilms on thermoplastic orthodontic appliances in 'fast' and 'slow' plaque formers. Eur J Orthod. 33:577–83. DOI: 10.1093/ejo/cjq126. PMID: 21187528. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=80053464335&origin=inward.
2. Naranjo AA, Triviño ML, Jaramillo A, Betancourth M, Botero JE. 2006; Changes in the subgingival microbiota and periodontal parameters before and 3 months after bracket placement. Am J Orthod Dentofacial Orthop. 130:275.e17–22. DOI: 10.1016/j.ajodo.2005.10.022. PMID: 16979483. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=33748440367&origin=inward.
3. Lucchese A, Gherlone E. 2013; Prevalence of white-spot lesions before and during orthodontic treatment with fixed appliances. Eur J Orthod. 35:664–8. DOI: 10.1093/ejo/cjs070. PMID: 23045306. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84885082210&origin=inward.
4. Eliades T, Eliades G, Brantley WA. 1995; Microbial attachment on orthodontic appliances: I. Wettability and early pellicle formation on bracket materials. Am J Orthod Dentofacial Orthop. 108:351–60. DOI: 10.1016/S0889-5406(95)70032-3. PMID: 7572846. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=0029393618&origin=inward.
5. Lim BS, Lee SJ, Lee JW, Ahn SJ. 2008; Quantitative analysis of adhesion of cariogenic streptococci to orthodontic raw materials. Am J Orthod Dentofacial Orthop. 133:882–8. DOI: 10.1016/j.ajodo.2006.07.027. PMID: 18538253. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=44449096172&origin=inward.
6. Komori R, Sato T, Takano-Yamamoto T, Takahashi N. 2012; Microbial composition of dental plaque microflora on first molars with orthodontic bands and brackets, and the acidogenic potential of these bacteria. J Oral Biosci. 54:107–12. DOI: 10.1016/j.job.2012.01.009. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84867575089&origin=inward.
7. Kolenbrander PE, Palmer RJ Jr, Periasamy S, Jakubovics NS. 2010; Oral multispecies biofilm development and the key role of cell-cell distance. Nat Rev Microbiol. 8:471–80. DOI: 10.1038/nrmicro2381. PMID: 20514044. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=77953620116&origin=inward.
8. Haririan H, Andrukhov O, Bertl K, Lettner S, Kierstein S, Moritz A, et al. 2014; Microbial analysis of subgingival plaque samples compared to that of whole saliva in patients with periodontitis. J Periodontol. 85:819–28. DOI: 10.1902/jop.2013.130306. PMID: 24144271. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84903436418&origin=inward.
9. Williams DL, Bloebaum RD. 2010; Observing the biofilm matrix of Staphylococcus epidermidis ATCC 35984 grown using the CDC biofilm reactor. Microsc Microanal. 16:143–52. DOI: 10.1017/S143192760999136X. PMID: 20205969. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=77952542903&origin=inward.
10. Good RJ. 1992; Contact angle, wetting, and adhesion: a critical review. J Adhes Sci Technol. 6:1269–302. DOI: 10.1163/156856192X00629. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84953125819&origin=inward.
11. McBain AJ, Sissons C, Ledder RG, Sreenivasan PK, De Vizio W, Gilbert P. 2005; Development and characterization of a simple perfused oral microcosm. J Appl Microbiol. 98:624–34. DOI: 10.1111/j.1365-2672.2004.02483.x. PMID: 15715865. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=14844303548&origin=inward.
12. Viana CS, Maske TT, Signori C, VAN DE Sande FH, Oliveira EF, Cenci MS. 2021; Influence of caries activity and number of saliva donors: mineral and microbiological responses in a microcosm biofilm model. J Appl Oral Sci. 29:e20200778. DOI: 10.1590/1678-7757-2020-0778. PMID: 34495103. PMCID: PMC8425900. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85114852064&origin=inward.
13. Shu M, Browngardt CM, Chen YY, Burne RA. 2003; Role of urease enzymes in stability of a 10-species oral biofilm consortium cultivated in a constant-depth film fermenter. Infect Immun. 71:7188–92. DOI: 10.1128/IAI.71.12.7188-7192.2003. PMID: 14638814. PMCID: PMC308945. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=0345283039&origin=inward.
14. Donlan RM, Piede JA, Heyes CD, Sanii L, Murga R, Edmonds P, et al. 2004; Model system for growing and quantifying Streptococcus pneumoniae biofilms in situ and in real time. Appl Environ Microbiol. 70:4980–8. DOI: 10.1128/AEM.70.8.4980-4988.2004. PMID: 15294838. PMCID: PMC492445. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=4143132359&origin=inward.
15. An JS, Kim K, Cho S, Lim BS, Ahn SJ. 2017; Compositional differences in multi-species biofilms formed on various orthodontic adhesives. Eur J Orthod. 39:528–33. DOI: 10.1093/ejo/cjw089. PMID: 28339597. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85028725118&origin=inward.
16. Rudney JD, Chen R, Lenton P, Li J, Li Y, Jones RS, et al. 2012; A reproducible oral microcosm biofilm model for testing dental materials. J Appl Microbiol. 113:1540–53. DOI: 10.1111/j.1365-2672.2012.05439.x. PMID: 22925110. PMCID: PMC3501590. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84869872306&origin=inward.
17. Park JW, An JS, Lim WH, Lim BS, Ahn SJ. 2019; Microbial changes in biofilms on composite resins with different surface roughness: an in vitro study with a multispecies biofilm model. J Prosthet Dent. 122:493.e1–493.e8. DOI: 10.1016/j.prosdent.2019.08.009. PMID: 31648793. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85073918757&origin=inward.
18. Yin JL, Shackel NA, Zekry A, McGuinness PH, Richards C, Putten KV, et al. 2001; Real-time reverse transcriptase-polymerase chain reaction (RT-PCR) for measurement of cytokine and growth factor mRNA expression with fluorogenic probes or SYBR Green I. Immunol Cell Biol. 79:213–21. DOI: 10.1046/j.1440-1711.2001.01002.x. PMID: 11380673. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=0034993341&origin=inward.
19. Fournier A, Payant L, Bouclin R. 1998; Adherence of Streptococcus mutans to orthodontic brackets. Am J Orthod Dentofacial Orthop. 114:414–7. DOI: 10.1016/S0889-5406(98)70186-6. PMID: 9790325. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=0032184822&origin=inward.
20. Ahn SJ, Kho HS, Lee SW, Nahm DS. 2002; Roles of salivary proteins in the adherence of oral streptococci to various orthodontic brackets. J Dent Res. 81:411–5. DOI: 10.1177/154405910208100611. PMID: 12097434. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=0035983426&origin=inward.
21. Lindel ID, Elter C, Heuer W, Heidenblut T, Stiesch M, Schwestka-Polly R, et al. 2011; Comparative analysis of long-term biofilm formation on metal and ceramic brackets. Angle Orthod. 81:907–14. DOI: 10.2319/102210-616.1. PMID: 21542722. PMCID: PMC8916195. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=80052482361&origin=inward.
22. Watanabe I, Watanabe E. 2003; Surface changes induced by fluoride prophylactic agents on titanium-based orthodontic wires. Am J Orthod Dentofacial Orthop. 123:653–6. DOI: 10.1016/S0889-5406(03)00197-5. PMID: 12806345. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=0038507442&origin=inward.
23. McBain AJ. 2009; In vitro biofilm models: an overview. Adv Appl Microbiol. 69:99–132. DOI: 10.1016/S0065-2164(09)69004-3. PMID: 19729092. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=69249171333&origin=inward.
24. Nyvad B, Fejerskov O. 1987; Scanning electron microscopy of early microbial colonization of human enamel and root surfaces in vivo. Scand J Dent Res. 95:287–96. DOI: 10.1111/j.1600-0722.1987.tb01844.x. PMID: 3476984. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84989593861&origin=inward.
25. Quirynen M, Marechal M, Busscher HJ, Weerkamp AH, Darius PL, van Steenberghe D. 1990; The influence of surface free energy and surface roughness on early plaque formation. An in vivo study in man. J Clin Periodontol. 17:138–44. DOI: 10.1111/j.1600-051X.1990.tb01077.x. PMID: 2319000. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=0025054981&origin=inward.
26. Busscher HJ, van Pelt AWJ, de Boer P, de Jong HP, Arends J. 1984; The effect of surface roughening of polymers on measured contact angles of liquids. Colloids Surf. 9:319–31. DOI: 10.1016/0166-6622(84)80175-4. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=0021423982&origin=inward.
27. An YH, Friedman RJ. 1998; Concise review of mechanisms of bacterial adhesion to biomaterial surfaces. J Biomed Mater Res. 43:338–48. DOI: 10.1002/(SICI)1097-4636(199823)43:3<338::AID-JBM16>3.0.CO;2-B. PMID: 9730073.
28. ten Cate JM. 2006; Biofilms, a new approach to the microbiology of dental plaque. Odontology. 94:1–9. DOI: 10.1007/s10266-006-0063-3. PMID: 16998612. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=33749059765&origin=inward.
29. Bollen CM, Lambrechts P, Quirynen M. 1997; Comparison of surface roughness of oral hard materials to the threshold surface roughness for bacterial plaque retention: a review of the literature. Dent Mater. 13:258–69. DOI: 10.1016/S0109-5641(97)80038-3. PMID: 11696906. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=0031173323&origin=inward.
30. Simonson LG, Reiher DA. 1981; Effect of human saliva and various compounds on the adsorption of the bacterium Streptococcus mutans to hydroxyapatite. Arch Oral Biol. 26:143–6. DOI: 10.1016/0003-9969(81)90085-6. PMID: 6944026. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=0019419122&origin=inward.
31. Suzuki N, Yoshida A, Nakano Y. 2005; Quantitative analysis of multi-species oral biofilms by TaqMan Real-Time PCR. Clin Med Res. 3:176–85. DOI: 10.3121/cmr.3.3.176. PMID: 16160072. PMCID: PMC1237159. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=27744600265&origin=inward.
32. Ahn SJ, Burne RA. 2007; Effects of oxygen on biofilm formation and the AtlA autolysin of Streptococcus mutans. J Bacteriol. 189:6293–302. DOI: 10.1128/JB.00546-07. PMID: 17616606. PMCID: PMC1951938. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=34548480696&origin=inward.
33. Kirthiga M, Murugan M, Saikia A, Kirubakaran R. 2019; Risk factors for early childhood caries: a systematic review and meta-analysis of case control and cohort studies. Pediatr Dent. 41:95–112. PMID: 30992106. PMCID: PMC7100045. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85064973342&origin=inward.
34. Wang BY, Lu T, Cai Q, Ho MH, Sheng S, Meng HW, et al. 2021; Potential microbiological risk factors associated with periodontitis and periodontal health disparities. Front Cell Infect Microbiol. 11:789919. DOI: 10.3389/fcimb.2021.789919. PMID: 34869082. PMCID: PMC8637773. PMID: 940959c0bbb24e50acad281cfc84c307. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85120677662&origin=inward.
Figure 1
Scanning electron microscope images of materials used in this study. The images were taken at 3,000× magnification. A, Monocrystalline alumina ceramic. B, Stainless steel metal. C, Polycarbonate plastic. D, Hydroxyapatite.
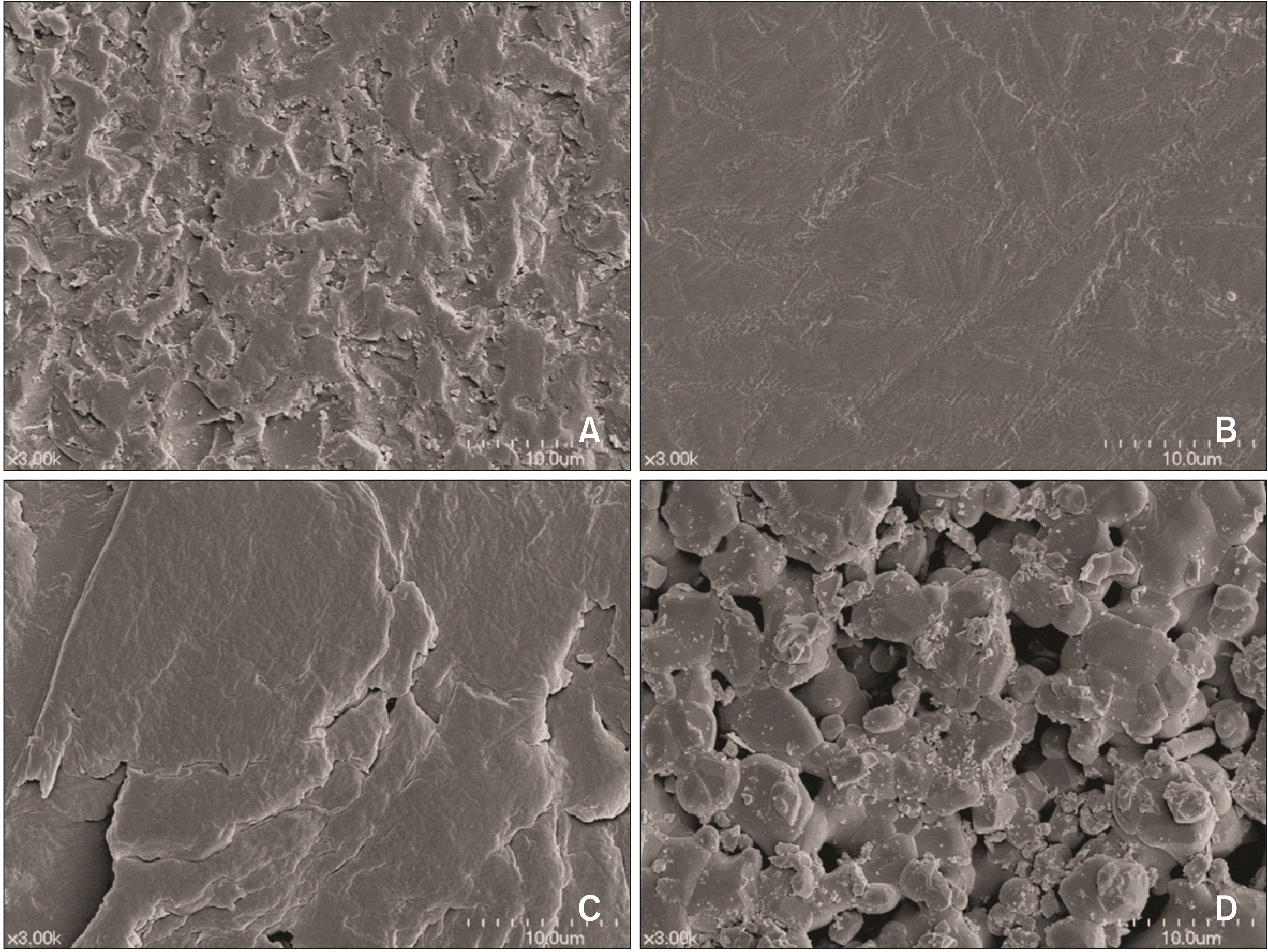
Table 1
Primers and cycling conditions used in this study
Table 2
Surface roughness and water contact angle of bracket materials used in this study
| Surface characteristics | CR | SS | PL | HA | Significance | p-value† |
|---|---|---|---|---|---|---|
| Surface roughness (μm) | 0.61 ± 0.24 | 0.55 ± 0.20 | 0.85 ± 0.27 | 1.26 ± 0.13 | CR = SS < PL < HA | < 0.001*** |
| Water contact angle (°) | 51.16 ± 26.06 | 85.39 ± 10.5 | 96.49 ± 7.89 | 42.69 ± 16.5 | CR = HA < SS < PL | < 0.001*** |
Table 3
Bacterial adhesion level with respect to material type and incubation time
| Level of bacterial adhesion | Day 1 | Day 4 | Significance (p-value)† | |
|---|---|---|---|---|
| Time | Material | |||
| Total bacteria adhesion level (Log10/cm2)‡ | ||||
| CR | 11.57 ± 0.35 | 12.29 ± 0.43 |
Day 1 < Day 4 (< 0.001***) |
CR = SS = PL < HA (< 0.001***) |
| SS | 11.66 ± 0.19 | 12.29 ± 0.51 | ||
| PL | 11.65 ± 0.17 | 12.19 ± 0.46 | ||
| HA | 11.80 ± 0.19 | 12.45 ± 0.39 | ||
| Streptococcus mutans adhesion level (Log10/cm2)‡ | ||||
| CR | 6.04 ± 0.49 | 6.43 ± 0.83 |
Day 1 < Day 4 (< 0.001***) |
SS = PL < CR = HA (< 0.001***) |
| SS | 5.72 ± 0.44 | 6.16 ± 0.87 | ||
| PL | 5.85 ± 0.35 | 6.22 ± 0.69 | ||
| HA | 6.39 ± 0.46 | 6.76 ± 0.85 | ||
| Porphyromonas gingivalis adhesion level (Log10/cm2)‡ | ||||
| CR | 5.85 ± 0.39 | 5.54 ± 0.52 |
Day 1 > Day 4 (< 0.01**) |
SS < CR = HA (< 0.01**) |
| SS | 5.46 ± 0.48 | 5.32 ± 0.48 | ||
| PL | 5.66 ± 0.46 | 5.51 ± 0.36 | ||
| HA | 5.84 ± 0.65 | 5.53 ± 0.54 | ||




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download