MATERIALS AND METHODS
Patients and specimens
PANAMutyper assay
NGS analysis
Statistical analysis
RESULTS
Prevalence of EGFR mutations according to the PANAMutyper test
Fig. 1
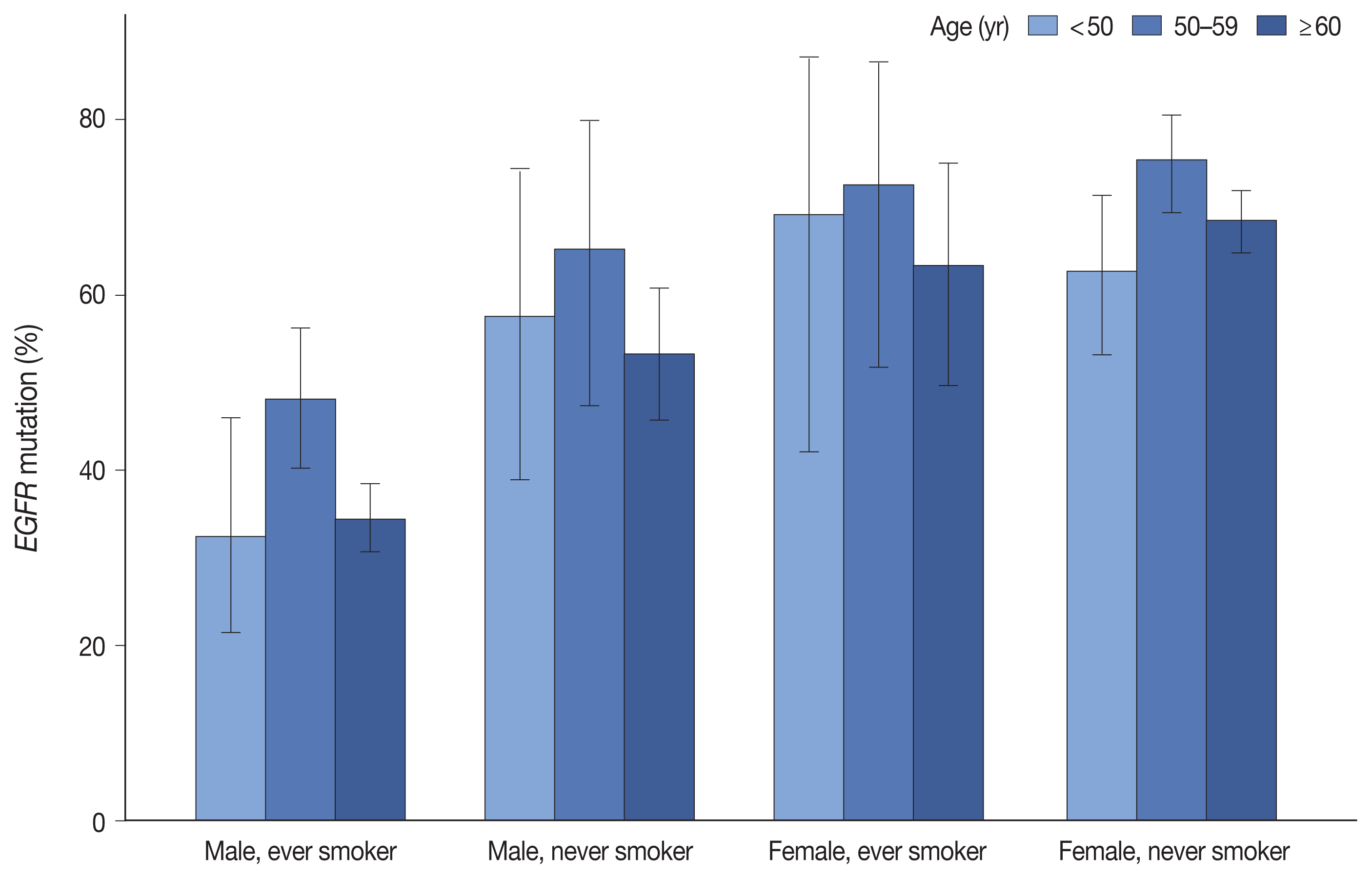
Table 1
| Characteristic | Examined No. | EGFR mutation | p-value |
|---|---|---|---|
| Total | 2,088 (100) | 1,162 (55.7) | |
| Age (yr) | |||
| < 40 | 36 (1.7) | 17 (47.2) | .001 |
| 40–49 | 161 (7.7) | 90 (55.9) | |
| 50–59 | 427 (20.5) | 277 (64.9) | |
| 60–69 | 673 (32.2) | 370 (55.0) | |
| 70–79 | 638 (30.6) | 334 (52.4) | |
| ≥ 80 | 153 (7.3) | 74 (48.4) | |
| Sex | |||
| Male | 1,005 (48.1) | 412 (41.0) | < .001 |
| Female | 1,083 (51.9) | 750 (69.3) | |
| Smoking statusa | |||
| Never | 1,205 (57.7) | 808 (67.1) | < .001 |
| Ever | 873 (41.8) | 349 (40.0) | |
| Sex and smoking statusa | |||
| Male, never smoker | 216 (10.3) | 120 (55.6) | < .001 |
| Male, ever smoker | 786 (37.6) | 291 (37.0) | |
| Female, never smoker | 989 (47.4) | 688 (69.6) | |
| Female, ever smoker | 87 (4.2) | 58 (66.7) | |
| Specimen type | |||
| Resection | 1,300 (62.3) | 773 (59.5) | < .001 |
| Primary lesion | 1,261 (60.4) | 746 (59.2) | |
| Metastatic lesion | 39 (1.9) | 27 (69.2) | |
| Biopsy | 776 (37.2) | 385 (49.6) | |
| Primary lesion | 463 (22.2) | 241 (52.1) | |
| PCNB | 345 (16.5) | 179 (51.9) | |
| Bronchoscopic biopsy | 117 (5.6) | 62 (53.0) | |
| Thoracoscopic biopsy | 1 (0) | 0 | |
| Metastatic lesion | 313 (15.0) | 144 (46.0) | |
| PCNB (other than LN) | 52 (2.5) | 29 (55.8) | |
| PCNB (LN) | 75 (3.6) | 32 (42.7) | |
| EBUS-TBNA biopsy (mediastinal LN) | 154 (7.4) | 64 (41.6) | |
| Thoracoscopic biopsy | 29 (1.4) | 18 (62.1) | |
| Other biopsies | 3 (0.1) | 1 (33.3) | |
| Cytology | 12 (0.6) | 4 (33.3) | |
| EBUS-TBNA | 4 (0.2) | 0 | |
| Pleural fluid | 8 (0.4) | 4 (50.0) | |
| Histologic subtypeb | |||
| Adenocarcinoma in situ | 16 (1.3) | 5 (31.3) | < .001 |
| Minimally invasive adenocarcinoma | 145 (11.5) | 92 (63.4) | |
| Lepidic adenocarcinoma | 55 (4.4) | 36 (65.5) | |
| Acinar adenocarcinoma | 404 (32.0) | 279 (69.1) | |
| Papillary adenocarcinoma | 348 (27.6) | 244 (70.1) | |
| Micropapillary adenocarcinoma | 52 (4.1) | 37 (71.2) | |
| Solid adenocarcinoma | 145 (11.5) | 48 (33.1) | |
| Invasive mucinous adenocarcinomac | 91 (7.2) | 5 (5.5) | |
| Colloid adenocarcinoma | 2 (0.2) | 0 | |
| Enteric-type adenocarcinoma | 3 (0.2) | 0 | |
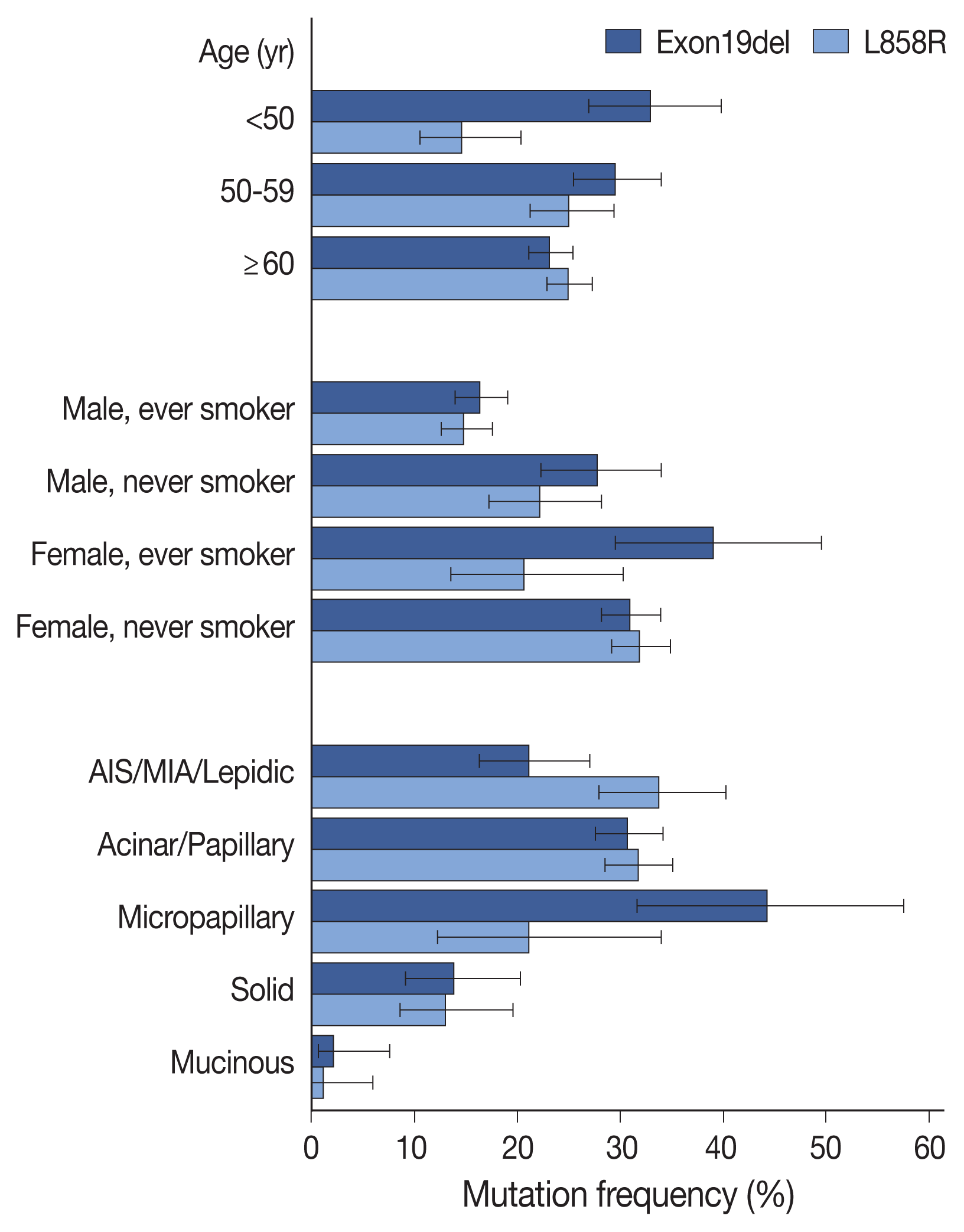
EGFR mutation subtypes
Table 2
| Total occurrence | Positive rate (%) | Relative proportion (%) | Form of mutation, n (%) | ||
|---|---|---|---|---|---|
|
|
|||||
| Singlet | Compound | ||||
| G719X | 49 | 2.3 | 4.2 | 31 (63.3) | 18 (36.7) |
| Exon19del | 530 | 25.4 | 45.7 | 521 (98.3) | 9 (1.7) |
| T790Ma | 12 | 0.6 | 1.0 | 3 (25.0) | 9 (75.0) |
| S768I | 12 | 0.6 | 1.0 | 3 (25.0) | 9 (75.0) |
| Exon20ins | 70 | 3.4 | 6.0 | 64 (91.4) | 6 (8.6) |
| L858R | 502 | 24.0 | 43.3 | 487 (97.0) | 15 (3.0) |
| L861Q | 19 | 0.9 | 1.6 | 15 (78.9) | 4 (21.1) |
Fig. 2
Repetitive tests
Comparison of PANAMutyper and NGS results for 73 patients
Fig. 3
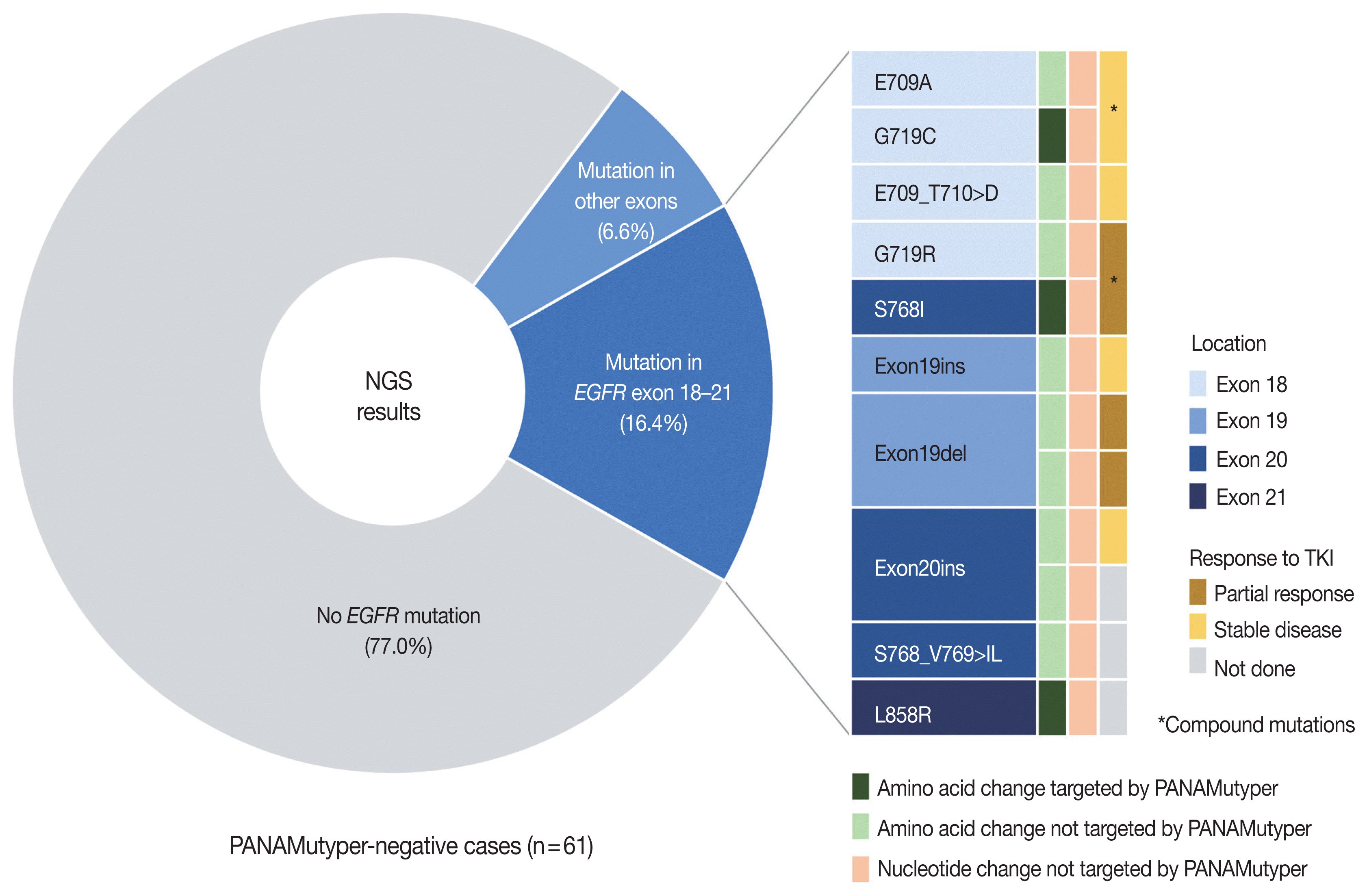
Table 3
| Case | Sex | Age (yr) | PANA results | NGS results | PANA-NGS samples | Sample type (PANA vs. NGS) | TKI treatment | Best response (TTD, mo) | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|||||||||||
| Exon | Nucleotide change | Amino acid change | VAF (%) | ||||||||
| N01 | M | 55 | Not identified |
18 18 |
c.2126A > C c.2154_2155delGGinsTT |
p.E709A p.G719C |
32.1 31.0 |
Same | Lung, PCNB | Afatinib | SD (32) |
| N02 | M | 55 | Not identified | 18 | c.2127_2129delAAC | p.E709_T710delinsD | 68.5 | Different | Lung, lobectomy (different block) | Afatinib | SD (15) |
| N03 | M | 54 | Not identified |
18 20 |
c.2155G > C c.2303_2304delGCinsTT |
p.G719R p.S768I |
27.5 33.7 |
Same | Lung, PCNB |
Erlotinib Afatinib Osimertinib |
PR (11) SD (7) PD (ND, 2) |
| N04 | M | 68 | Not identified | 19 | c.2214_2231dupTAAAATTCCCGTCGCTAT | p.I744_K745insKIPVAI | 33.7 | Different | Lung, lobectomy (different block) | Erlotinib | SD (24) |
| N05 | F | 34 | Not identified | 19 | c.2239_2253delTTAAGAGAAGCAACAinsAAC | p.L747_T751delinsN | 47.8 | Different | Lung, lobectomy (different block) | Erlotinib | PR (NR, 24+) |
| N06 | F | 50 | Not identified |
19 26 |
c.2253_2276delATCTCCGAAAGCCAACAAGGAAATinsTTCCGC c.3143C > T |
p.P753_I759delinsA p.A1048V |
10.8 50.0 |
Same | LN, PCNB | Erlotinib | PR (4) |
| N07 | F | 59 | Not identified | 20 | c.2284-5_2290dupTCCAGGAAGCCT | p.A763_Y764insFQEA | 20.2 | Same | LN, PCNB | Amivantamab | SD (NR, 9+) |
| N08 | M | 67 | Not identified |
20 26 |
c.2303_2305delGCGinsTCT c.3143C > T |
p.S768_V769 > IL p.A1048V |
51.7 60.7 |
Same | Lung, PCNB | Erlotiniba | PR (22)a |
| N09 | F | 71 | Not identified | 20 | c.2311_2319dupAACCCCCAC | p.N771_H773dup | 13.0 | Same | Lung, PCNB | Not done | - |
| N10 | M | 54 | Not identified | 21 | c.2573_2574delTGinsGC | p.L858R | 16.5 | Different | Peritoneum, excision (different lesion) | Not done | - |
| N11 | F | 75 | Not identified | 4 | c.500T > C | p.I167T | 3.8 | Same | Lung, PCNB | Not done | - |
| N12 | M | 68 | Not identified | 11 | c.1282G > A | p.G428S | 2.3 | Same | Lung, PCNB | Not done | - |
| N13 | M | 55 | Not identified | 26 | c.3143C > T | p.A1048V | 44.0 | Different | LN, EBUS-TBNA Bx (different node) | Not done | - |
| N14 | F | 49 | Not identified | 26 | c.3143C > T | p.A1048V | 49.9 | Same | Lung, PCNB | Not done | - |
| N15 | F | 58 | G719X |
18 18 |
c.2125G > A c.2156G > C |
p.E709K p.G719A |
42.2 41.0 |
Same | LN, PCNB |
Erlotiniba Afatinib |
SD (6)a N/A (ND, 4) |
| N16 | F | 36 | Exon19del |
18 19 |
c.2170G > A c.2237_2255delAATTAAGAGAAGCAACATCinsT |
p.G724S p.E746_S752delinsV |
9.8 32.7 |
Same | Pleura, VATS biopsy |
Gefitiniba Erlotinib |
SD (12.5)a SD (9) |
| N17 | M | 66 | Exon19del | 17 | c.1996C > T | p.L666F | 45.9 | Same | Lung, PCNB | Erlotinib | SD (ND, 2) |
| N18 | F | 50 | Exon19del | Not identified | Different | LN, EBUS-TBNA Bx vs. pleural fluid |
Erlotiniba Erlotinib |
PD (1)a PD (1) |
|||
EGFR, epidermal growth factor receptor; NGS, next-generation sequencing; PANA, PANAMutyper; VAF, variant allele frequency; TKI, tyrosine kinase inhibitor; TTD, time-to-treatment discontinuation; M, male; F, female; PCNB, percutaneous needle biopsy; SD, stable disease; PR, partial response; PD, progressive disease; ND, not detected; NR, not reached; LN, lymph node; EBUS-TBNA, endobronchial ultrasound-transbronchial needle aspiration; Bx, biopsy; N/A, not available; VATS, video-assisted thoracoscopic surgery.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download