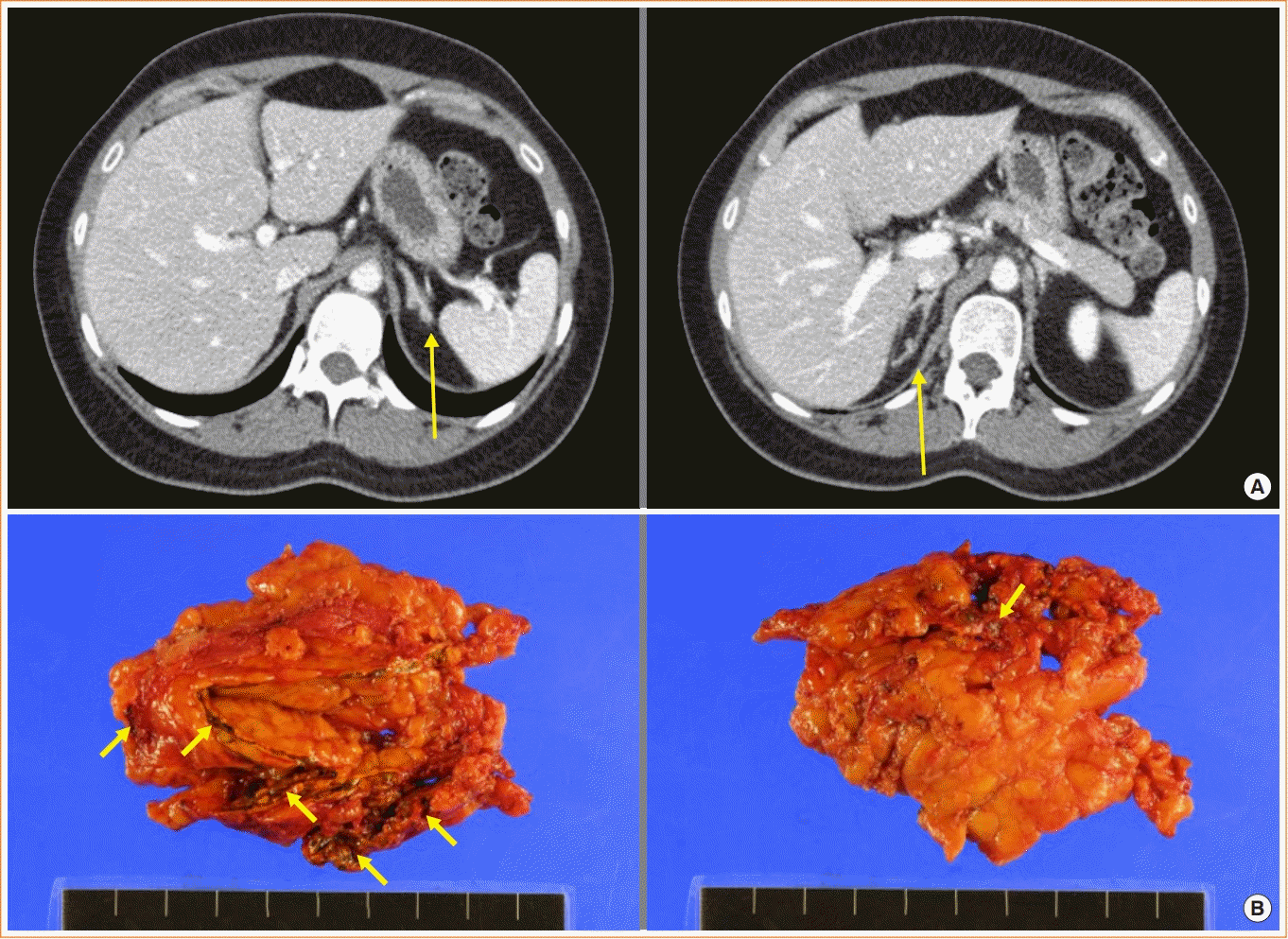
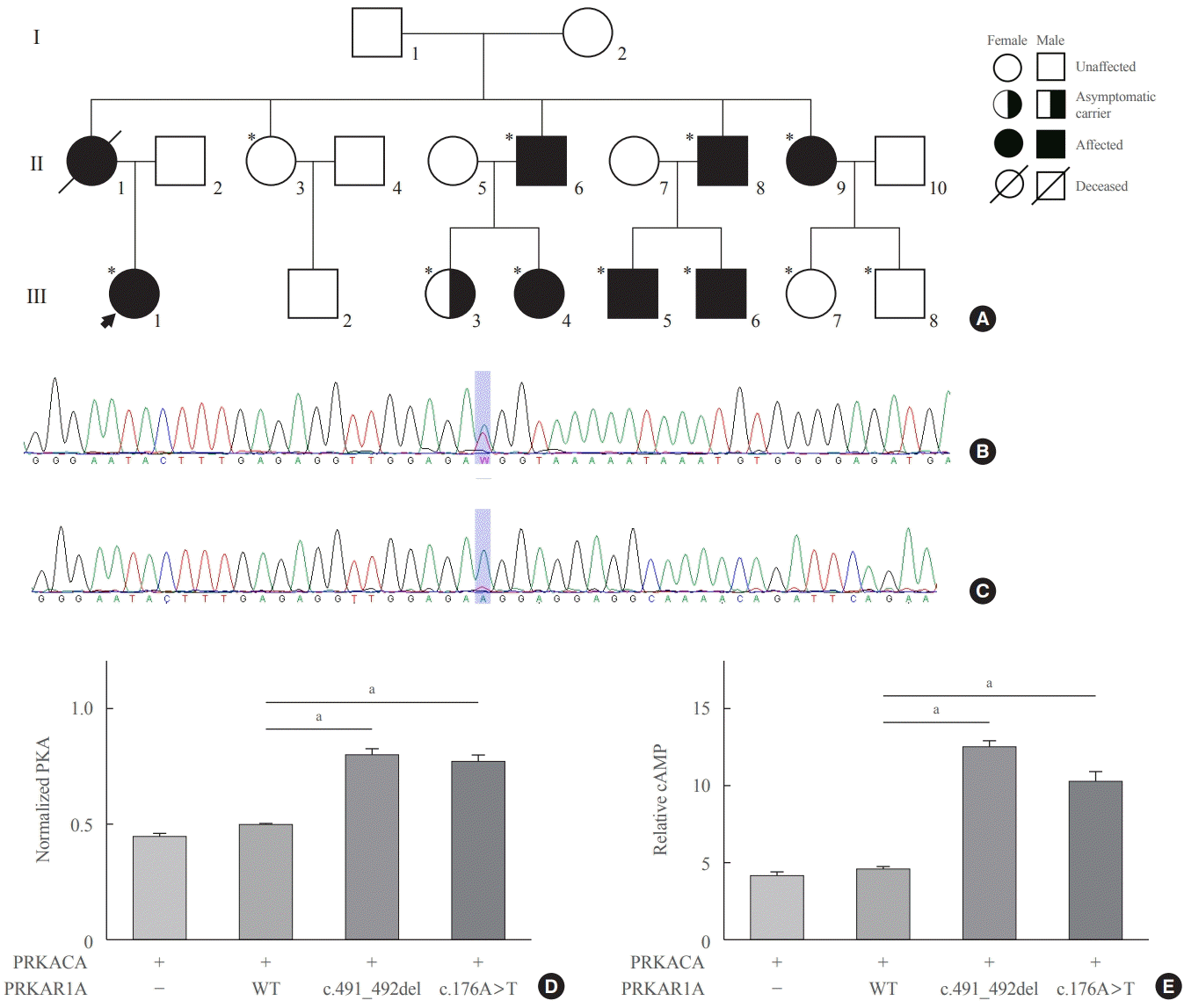
1. Carney JA, Gordon H, Carpenter PC, Shenoy BV, Go VL. The complex of myxomas, spotty pigmentation, and endocrine overactivity. Medicine (Baltimore). 1985; 64:270–83.

2. Bouys L, Bertherat J. Management of endocrine disease. Carney complex: clinical and genetic update 20 years after the identification of the CNC1 (PRKAR1A) gene. Eur J Endocrinol. 2021; 184:R99–109.

3. Salpea P, Stratakis CA. Carney complex and McCune Albright syndrome: an overview of clinical manifestations and human molecular genetics. Mol Cell Endocrinol. 2014; 386:85–91.

4. Kamilaris CD, Faucz FR, Voutetakis A, Stratakis CA. Carney complex. Exp Clin Endocrinol Diabetes. 2019; 127:156–64.

5. Correa R, Salpea P, Stratakis CA. Carney complex: an update. Eur J Endocrinol. 2015; 173:M85–97.

6. Zhang CD, Pichurin PN, Bobr A, Lyden ML, Young WF, Bancos I. Cushing syndrome: uncovering Carney complex due to novel PRKAR1A mutation. Endocrinol Diabetes Metab Case Rep. 2019; 2019:18–0150.

7. Steinhaus R, Proft S, Schuelke M, Cooper DN, Schwarz JM, Seelow D. MutationTaster2021. Nucleic Acids Res. 2021; 49(W1):W446–51.

8. Sim NL, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012; 40(Web Server issue):W452–7.

9. Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010; 7:248–9.

10. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015; 17:405–24.

11. Jian X, Boerwinkle E, Liu X. In silico prediction of splicealtering single nucleotide variants in the human genome. Nucleic Acids Res. 2014; 42:13534–44.

12. Bertherat J, Horvath A, Groussin L, Grabar S, Boikos S, Cazabat L, et al. Mutations in regulatory subunit type 1A of cyclic adenosine 5’-monophosphate-dependent protein kinase (PRKAR1A): phenotype analysis in 353 patients and 80 different genotypes. J Clin Endocrinol Metab. 2009; 94:2085–91.
13. Espiard S, Vantyghem MC, Assie G, Cardot-Bauters C, Raverot G, Brucker-Davis F, et al. Frequency and incidence of carney complex manifestations: a prospective multicenter study with a three-year follow-up. J Clin Endocrinol Metab. 2020; 105:dgaa002.

14. Horvath A, Bertherat J, Groussin L, Guillaud-Bataille M, Tsang K, Cazabat L, et al. Mutations and polymorphisms in the gene encoding regulatory subunit type 1-alpha of protein kinase A (PRKAR1A): an update. Hum Mutat. 2010; 31:369–79.

15. NICHD PRKAR1A Mutation Database. Regulatory Subunit type 1-alpha of protein kinase A (PKA): mutation database [Internet]. Bethesda: National Institutes of Health;2022. [cited 2022 Sep 13]. Available from:
https://prkar1a.nichd.nih.gov/.
16. Stratakis CA, Kirschner LS, Carney JA. Clinical and molecular features of the Carney complex: diagnostic criteria and recommendations for patient evaluation. J Clin Endocrinol Metab. 2001; 86:4041–6.