Abstract
Objectives
Automated systems for information extraction are becoming very useful due to the enormous scale of the existing literature and the increasing number of scientific articles published worldwide in the field of medicine. We aimed to develop an accessible method using the open-source platform KNIME to perform text mining (TM) on indexed publications. Material from scientific publications in the field of life sciences was obtained and integrated by mining information on hemolytic uremic syndrome (HUS) as a case study.
Methods
Text retrieved from Europe PubMed Central (PMC) was processed using specific KNIME nodes. The results were presented in the form of tables or graphical representations. Data could also be compared with those from other sources.
Results
By applying TM to the scientific literature on HUS as a case study, and by selecting various fields from scientific articles, it was possible to obtain a list of individual authors of publications, build bags of words and study their frequency and temporal use, discriminate topics (HUS vs. atypical HUS) in an unsupervised manner, and cross-reference information with a list of FDA-approved drugs.
Conclusions
Following the instructions in the tutorial, researchers without programming skills can successfully perform TM on the indexed scientific literature. This methodology, using KNIME, could become a useful tool for performing statistics, analyzing behaviors, following trends, and making forecast related to medical issues. The advantages of TM using KNIME include enabling the integration of scientific information, helping to carry out reviews, and optimizing the management of resources dedicated to basic and clinical research.
A widespread problem when analyzing publications on biomedical topics is the exponential increase in the number of articles published each year. The use of automated systems to extract information from published articles has become a necessity [1,2]. Tools that facilitate the retrieval and articulation of digital information also make it possible to integrate “fragments of knowledge” into models that help manage complex problems and reduce costs in health prevention and the treatment of pathologies [3].
One of these tools is text mining (TM), which allows the examination and analysis of large collections of written resources, transforming the text used to represent language and the explicit knowledge into data to generate new information. According to Hotho et al. [4] there are three possible approaches to TM: information extraction, data mining, and knowledge discovery in databases.
In this tutorial, we show how to perform TM in medical articles in an accessible way that enables the discovery of non-explicit (often hidden) information structures and patterns through KNIME (https://www.knime.com/). The KNIME Analytics Platform is free, open-source software for creating visual workflows for data analytics and using nodes in successive steps, with the possibility of inspecting each partial result.
The data corpus in this tutorial comprised the publications indexed in Europe PubMed Central (ePMC, https://europepmc.org/). ePMC is an open scientific platform that provides access to a global collection of life science publications from reliable sources. ePMC was developed by the European Bioinformatics Institute (EMBL-EBI), a partner of PubMed Central, but it outnumbers PubMed Central by more than 5 million abstracts. ePMC also contains patents, NHS (National Health Service) guidelines, and agricultural records.
The methodology described in the present tutorial makes it possible to relate dispersed data and to present the data in a compact and clear manner, leading to a deeper understanding of several descriptors. It also detects fluctuations and trends and is capable of extracting implicit and hidden information and cross-referencing them with other sources of interest. With minor adjustments, this methodology also makes it possible to obtain statistical information about journals, authors, institutions, and countries involved in the research. When applied to the words used by authors, TM helps to detect undescribed associations between events and to cluster words thematically with unsupervised algorithms.
The procedure described in this study was tested and applied to the analysis of a database of more than 75,000 publications [5], using standard computers; its design enables it to work with even larger databases. Several workflows were initially designed to mine publications on hemolytic uremic syndrome (HUS) [6]. HUS is recognized as the most common cause of acute kidney failure in infants and young children, although it can also affect adolescents and adults. HUS is a clinical syndrome usually categorized as typical or atypical [7] and defined as the triad of microangiopathic hemolytic anemia, thrombocytopenia, and acute kidney injury [8]. Typical HUS, which is caused by Shiga toxin-producing Escherichia coli (STEC) infection and is therefore also called STEC-HUS, is the most frequent type of HUS; it is caused by ingestion of contaminated foodstuffs and through animal or person-to-person contact. Atypical HUS (aHUS) is associated with mutations or autoantibodies leading to dys-regulated complement activation or is secondary to a coexisting disease.
This tutorial presents the application of the TM-with-KNIME method for scientific articles on HUS published in 2020 and 2021 as a case study.
A standard computer with a modern processor, 16 GB RAM, 1 TB hard drive, and the Windows 10, Linux, or macOS operating system can be used. The KNIME Analytics Platform must be installed following the instructions at https://www.knime.com/installation. After the basic installation, specific extensions must be installed to run the TM workflows. As shown in Figure 1A, these extensions must be added from within KNIME by selecting “Install KNIME Extensions” from the File menu. A new installation window opens with the option to type in a keyword (Figure 1B). The keyword “text” is sufficient to display the KNIME Text-processing extension (English nodes are the default, but it is possible to select nodes to perform TM in other languages). This extension must be selected and installed. Similarly, by typing “vernalis” it is possible to install KNIME Community Extensions-Cheminformatics with the KNIME Vernalis nodes, and by typing “indexing,” one can install the KNIME Labs Extensions with the KNIME Indexing and Searching node.
The indexed information is retrieved from the ePMC site using the European PubMed Central Advanced Search KNIME node (Figure 1). It is recommended to perform an Advanced Search at https://europepmc.org/advancesearch using keywords and filters on the fields of interest. The resulting syntax in the Advanced Search windows may be copied and pasted into the General Query field in the node configuration window (Figure 1C). In the example, the syntax (“haemolytic uraemic syndrome” OR “hemolytic uremic syndrome”) was used to include two spellings of search terms for HUS publications. The years of publication were limited to 2020–2021. After the execution of this first node, the XPath node was used to select the information to work in, as exemplified by the paths shown in Figure 1D. Note that for the affiliation field, the string type CollectionCell must be specified. The Table Indexer node (Figure 1E) allows the user to select columns to index the information that will be consulted in the following Index Query node. This central node allows the user to obtain the necessary information for further processing. Changing the syntax of the query text makes it possible to evaluate publications per year, the most and least common journals chosen by the authors, the list of authors, the countries where the authors work, the number of publications per author, and so on (Figure 1F). It also allows the user to obtain the full text of the titles and the abstracts. Some examples of syntax in the query text in the KNIME Index Query node are shown in Table 1. Specifically, abstracts are used to obtain the linguistic corpus of interest. Statistics and corresponding graphs can be made in KNIME (using Value Counter, Sorter, Scatter Plot and other specific nodes) or by exporting the data to any statistical software.
To obtain the full list of authors, we recommend the workflow shown in Figure 2. After a specific index query, the authorString field constitutes the corpus to extract information by successive application of nodes.
The automatic clustering of topics and the retrieval of a bag of words are two powerful tools for analyzing a corpus consisting of all the abstracts of publications; they are counted, depending on each query, in hundreds, thousands or even millions of items. The Topic Extractor node is used for the automatic and unsupervised detection of topics and keywords after an index query and a preprocessing of the text of the corpus (as detailed in Figure 3, fork 1). Topic extraction is based on a simple parallel threaded implementation of latent Dirichlet allocation (LDA), following Newman et al. [9], with a sparse LDA sampling scheme and data structure from Yao et al. [10]. This technique uses the Machine Learning for Language Toolkit (MALLET) topic modeling library.
The above-mentioned text preprocessing technique can be used to create a bag of words from a set of documents through the Bag of Words Creator node (Figure 3, fork 2). The use of different nodes results in table presentations or graphically ordered representations (as clouds of words), as shown in Figure 3.
Thematic clustering in our example showed a clear differentiation between HUS (topics 2 and 3) and aHUS (topics 0, 1, and 4), as shown in both the table and word cloud outputs.
As noted, the corpus of abstracts contains valuable information, which can be cross-referenced with external data sources to separate and rank data of interest. The workflow in Figure 4 shows a cross-check between the abstracts of articles on HUS published in 2020 and 2021 and the list of the Food and Drug Administration (FDA) approved drugs (https://www.fda.gov/drugs/development-approval-processdrugs/drug-approvals-and-databases). This workflow makes it possible to detect mentions of some of these drugs in abstracts. In fact, the corpus could be cross-checked with any other list of interest in the same way.
The main objective of this work was to describe an accessible method designed to discover non-explicit information about structures and patterns in the fields of scientific articles indexed in ePMC. A description of the text mining nodes used in the tutorial is shown in Table 2. The proposed approach, which used KNIME workflows, allowed the linkage and analysis of scattered data, leading to a deeper understanding of the topic under study.
As described elsewhere in the literature, KNIME has been shown to be a powerful data analysis tool [11]. During the current coronavirus disease 2019 (COVID-19) pandemic, the KNIME platform has been used to map the research domains explored through clinical trials related to COVID-19. More than 3,000 clinical trials were analyzed using a wordcloud that helped to identify various scientific areas explored in COVID-19-related clinical studies [12].
KNIME has proven to be versatile and useful in different fields of knowledge besides medical research, as shown by research in areas as diverse as marketing [13], geosciences [14], and social issues [15].
TM of the scientific literature can be considered as a tool for human health research and is an invaluable aid for researchers engaged in writing a review on their specialized topic, saving efforts in the selection and analysis of relevant publications. The strategy presented in this tutorial could be applied directly to the study of almost any scientific topic in human health or the life sciences.
Although other KNIME workflows could be implemented for the analysis of the full text of papers, we believe that abstracts contain the main ideas of the research. The full text of the publication may contain redundant information that distracts from the focus of the analysis, as well as requiring a large amount of computational time, which is not always available to research teams around the world. In this tutorial, we present workflows that allow a large number of results to be analyzed in depth, without much difficulty and using standard computers.
We hope that the new strategies using TM could help improve prevention, research, and treatment of different diseases, optimizing budgetary decisions related to specific topics or the choice of thematic approaches, and thereby increasing efficiency in the use of resources.
Finally, the proposed KNIME workflows, which use different aspects of TM, should be seen as a contribution to imagining new ways of approaching scientific texts in a simple and accessible manner.
Acknowledgments
This work was supported by grant “Proyecto de Investigación de Unidades Ejecutoras (P-UE 2017) No. 22920170100041CO” from Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), Argentina and “UBACyT No. 20020170100733BA” from Universidad de Buenos Aires (UBA), Argentina. We gratefully acknowledge the support of NVIDIA Corporation with the donation of the Titan Xp GPU used for our research. We thank Dr. Gisela Di Giusto for her critical reading to improve the manuscript. We thank Mrs. Laura Toledo for her language revision.
References
1. Renganathan V. Text mining in biomedical domain with emphasis on document clustering. Healthc Inform Res. 2017; 23(3):141–6. https://doi.org/10.4258/hir.2017.23.3.141.

2. Tshitoyan V, Dagdelen J, Weston L, Dunn A, Rong Z, Kononova O, et al. Unsupervised word embeddings capture latent knowledge from materials science literature. Nature. 2019; 571(7763):95–8. https://doi.org/10.1038/s41586-019-1335-8.

3. Viceconti M, Hunter P. The virtual physiological human: ten years after. Annu Rev Biomed Eng. 2016; 18:103–23. https://doi.org/10.1146/annurev-bioeng-110915-114742.

4. Hotho A, Nurnberger A, Paaß G. A brief survey of text mining. LDV Forum. 2005; 20(1):19–62.
5. Dorr RA, Casal JJ, Toriano R. Minería de texto en publicaciones científicas con autores argentinos [Text mining in scientific publications with Argentine authors]. Medicina (B Aires). 2021; 81(2):214–23.
6. Dorr RA, Silberstein C, Ibarra C, Toriano R. Obtaining new information on hemolytic uremic syndrome by text mining. Medicina (B Aires). 2022; 82(4):513–24. PMID: 35904906.
7. Jokiranta TS. HUS and atypical HUS. Blood. 2017; 129(21):2847–56. https://doi.org/10.1182/blood-2016-11-709865.

8. Exeni RA, Fernandez-Brando RJ, Santiago AP, Fiorentino GA, Exeni AM, Ramos MV, et al. Pathogenic role of inflammatory response during Shiga toxin-associated hemolytic uremic syndrome (HUS). Pediatr Nephrol. 2018; 33(11):2057–71. https://doi.org/10.1007/s00467-017-3876-0.

9. Newman D, Asuncion A, Smyth P, Welling M. Distributed algorithms for topic models. J Mach Learn Res. 2009; 10:1801–28.
10. Yao L, Mimno D, McCallum A. Efficient methods for topic model inference on streaming document collections. In : Proceedings of the 15th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; 2009 Jun 28–Jul 1; Paris, France. p. 937–46. https://doi.org/10.1145/1557019.1557121.

11. Qundus JA, Peikert S, Paschke A. AI supported topic modeling using KNIME-workflows [Internet]. Ithaca (NY): arXiv.org;2021. [cited at 2022 May 2]. Available from: https://arxiv.org/abs/2104.09428.
12. Patel S, Patel A, Patel M, Shah U, Patel M, Solanki N, et al. Review and analysis of massively registered clinical trials of COVID-19 using the text mining approach. Rev Recent Clin Trials. 2021; 16(3):242–57. https://doi.org/10.2174/1574887115666201202110919.

13. Ordenes FV, Silipo R. Machine learning for marketing on the KNIME Hub: the development of a live repository for marketing applications. J Bus Res. 2021; 137:393–410. https://doi.org/10.1016/j.jbusres.2021.08.036.

14. Feltrin L. KNIME an open source solution for predictive analytics in the geosciences [software and data sets]. IEEE Geosci Remote Sens Mag. 2015; 3(4):28–38. https://doi.org/10.1109/MGRS.2015.2496160.

15. Vijayan R. Teaching and learning during the COVID-19 pandemic: a topic modeling study. Educ Sci. 2021; 11(7):347. https://doi.org/10.3390/educsci11070347.

Figure 1
Example of starting a workflow. (A) From the KNIME File menu, select “Install KNIME Extensions.” (B) To select the required text mining extension (KNIME Textprocessing), “text” can be typed in the text box. Similarly, the Vernalis KNIME Nodes and KNIME Indexing and Searching extensions must be installed. (C) The workflow starts with a search on the European PubMed Central (ePMC) site. Specific query terms should be typed in the General Query text box. In our example, the query terms were (“Haemolytic uraemic syndrome” OR “Hemolytic uremic syndrome”), corresponding to two different spellings for the same clinical syndrome. The years of publication were limited to 2020–2021. The Test Query button is used to check the number of hits. The node returns an XML document. (D) The XPath node allows selecting the fields of interest (see Column Name) from the XML document. (E) All the fields are indexed with the Table Indexer node. (F) The Index Query node creates a filtered data table, which is the input corpus for the following nodes. In the figure, only the articles published in 2020 are selected. Configuration windows C, D, E, and F are opened with a double left click on the node icon (blue arrow).
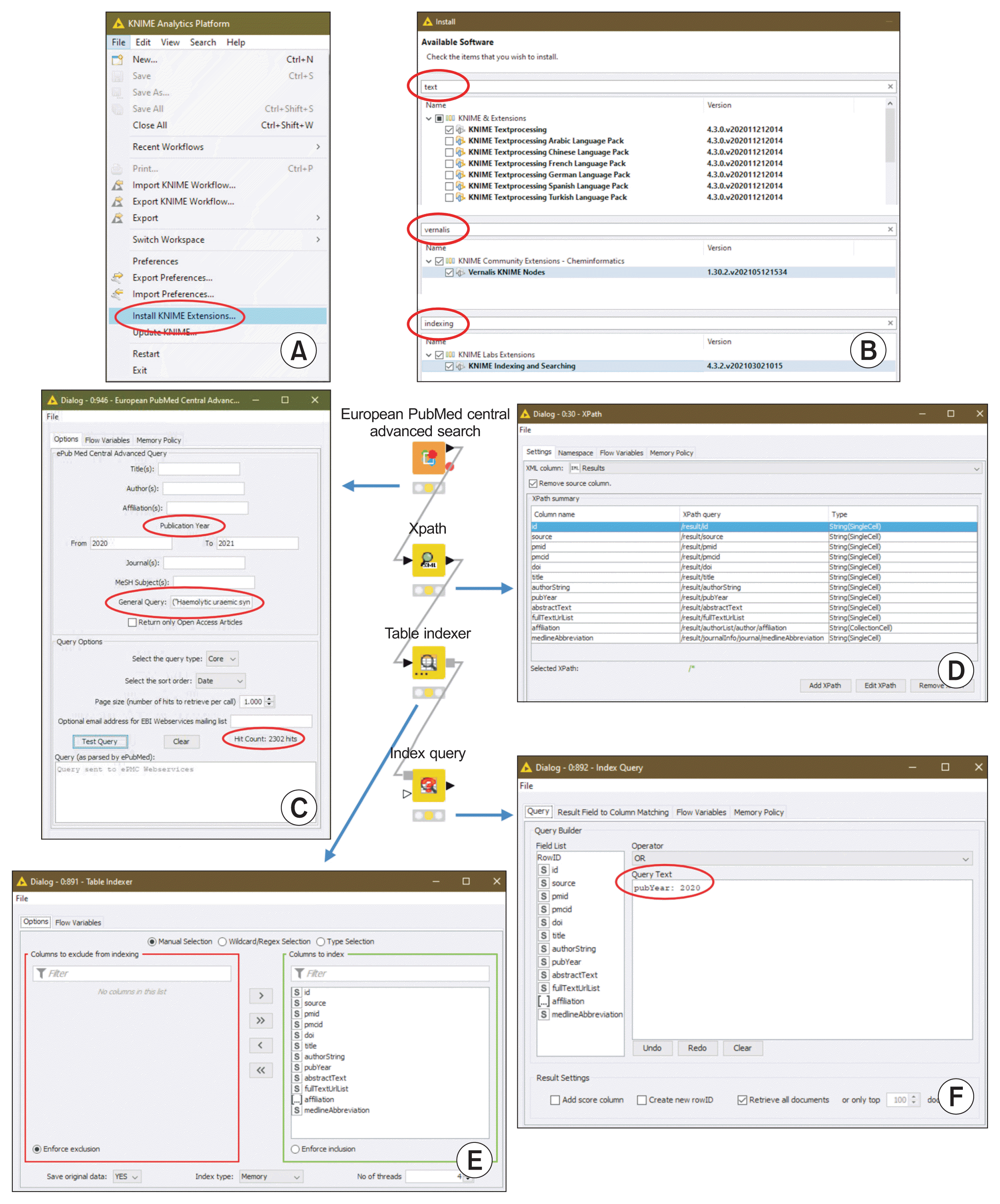
Figure 2
Example for obtaining a list of authors. The authorString table lists the authors signing the publication after running the Index Query node. All authors of a publication are in the same row (see entry detail). The transposition performed with the Transpose node and the splitting of a cell into its constituent components (Cell Splitter node) are used to obtain the individual list of authors (see output detail). The results of a node action can be viewed by opening a window with a right click on the node icon (green arrow). The orange arrow indicates a brief node description.
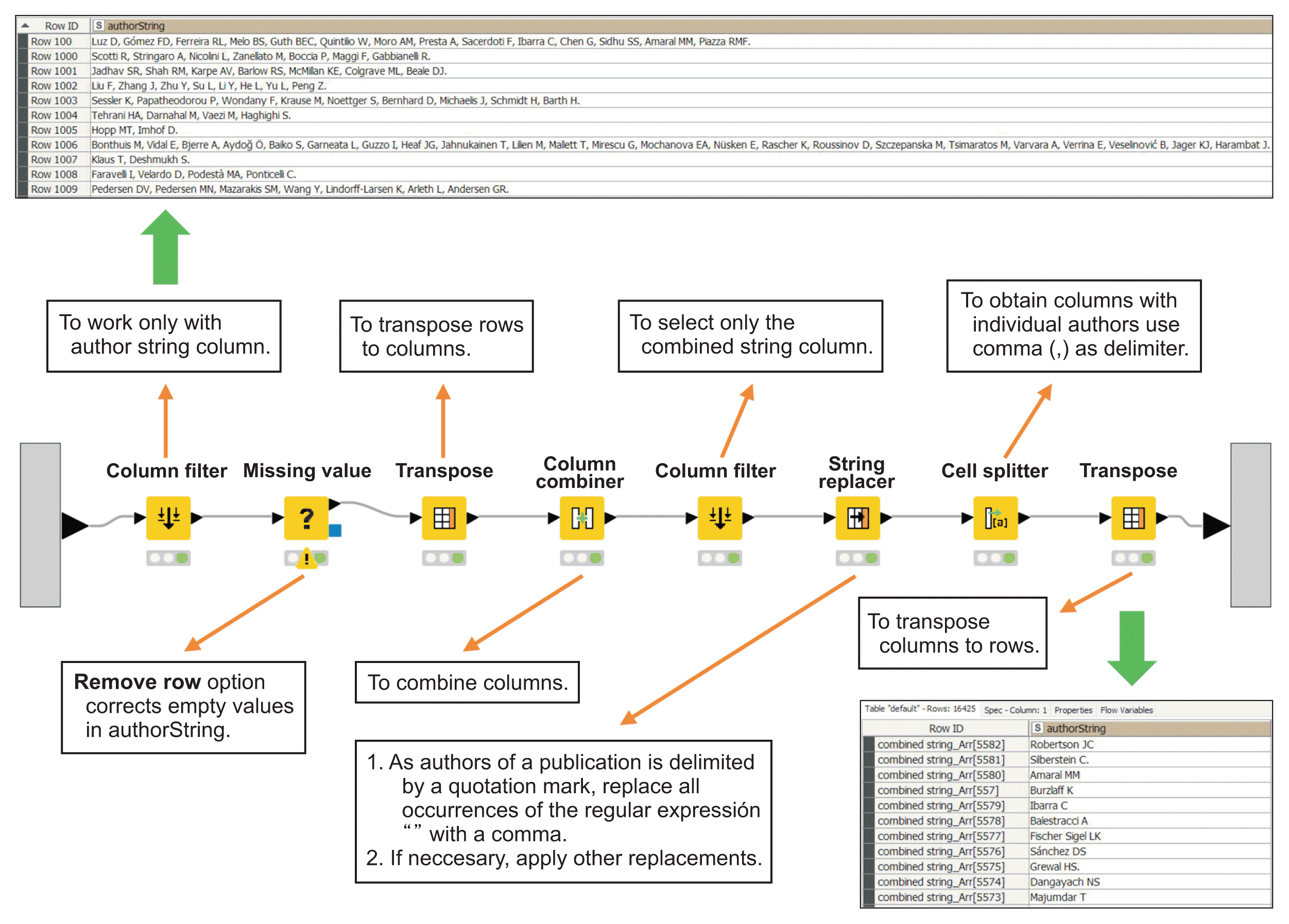
Figure 3
Example of automated and unsupervised detection of topics in abstracts about hemolytic uremic syndrome (HUS) and quantification of their characteristic words. The example shows the topics detected in publications on hemolytic uremic syndrome from 2020 to 2021 inclusive. A proposed text preprocessing method that facilitates subsequent analysis is also exemplified, eliminating characters and words without semantic importance, grouping by lemmatization and labeling the tokens. The result of topic detection (fork 1) is shown in tabular form but could also be presented in another graphical form. The word cloud (result of fork 2) represents the most abundant words in a bag of words; the larger its size, the higher its frequency of use. Words in a topic have the same color. Green arrow: output with right click, Orange arrow: brief node description.
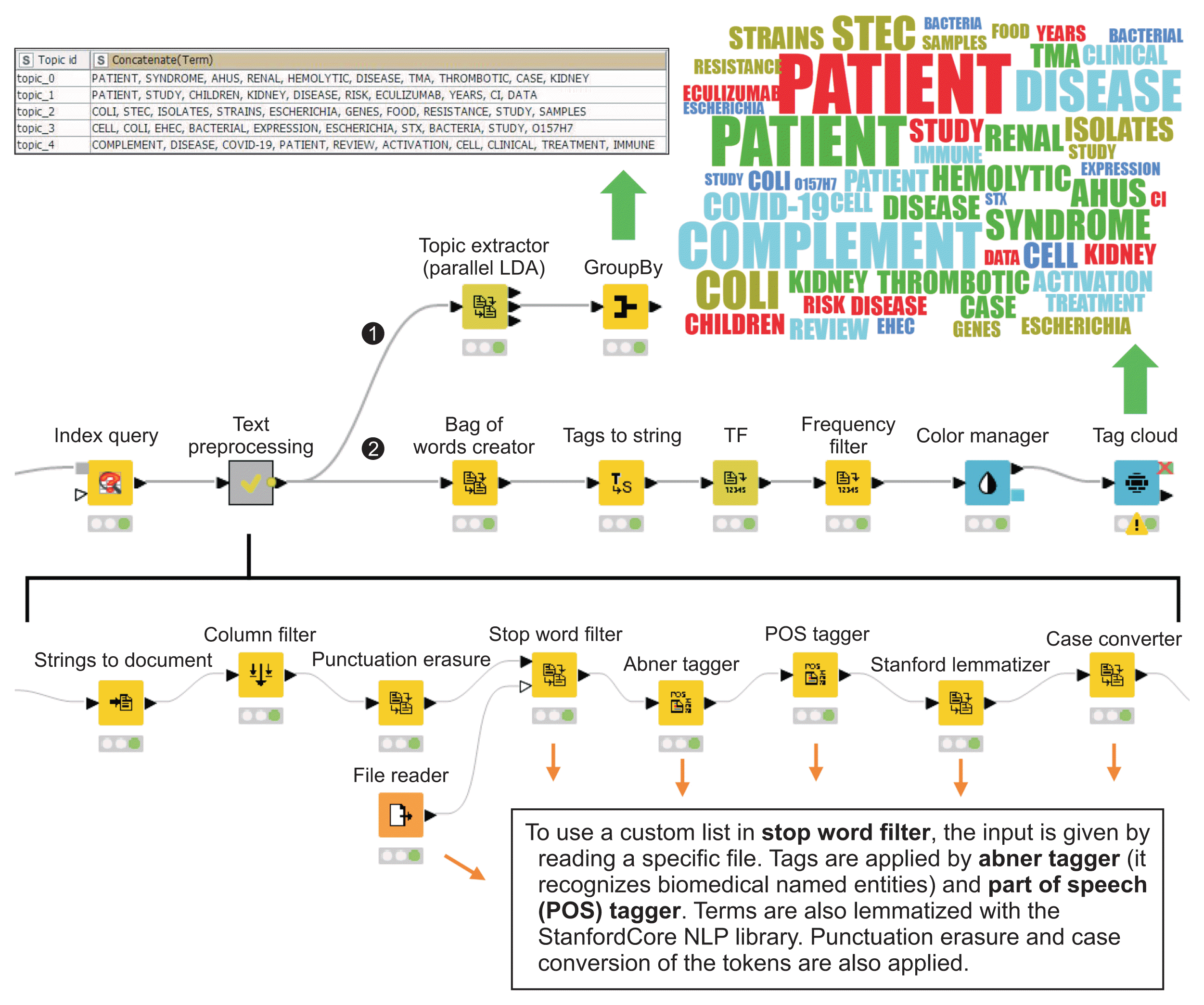
Figure 4
Workflow with cross-referencing. The input table (see details at the top) contains the terms to be identified in the corpus by means of the Cross Joiner node. Unrecognized terms are excluded by applying a filter in the Row Filter node. The output shows the number of times that each FDA-approved drug was found in abstracts (see details at the bottom). Green arrow: output with right click, Orange arrow: brief node description, FDA: Food and Drug Administration.
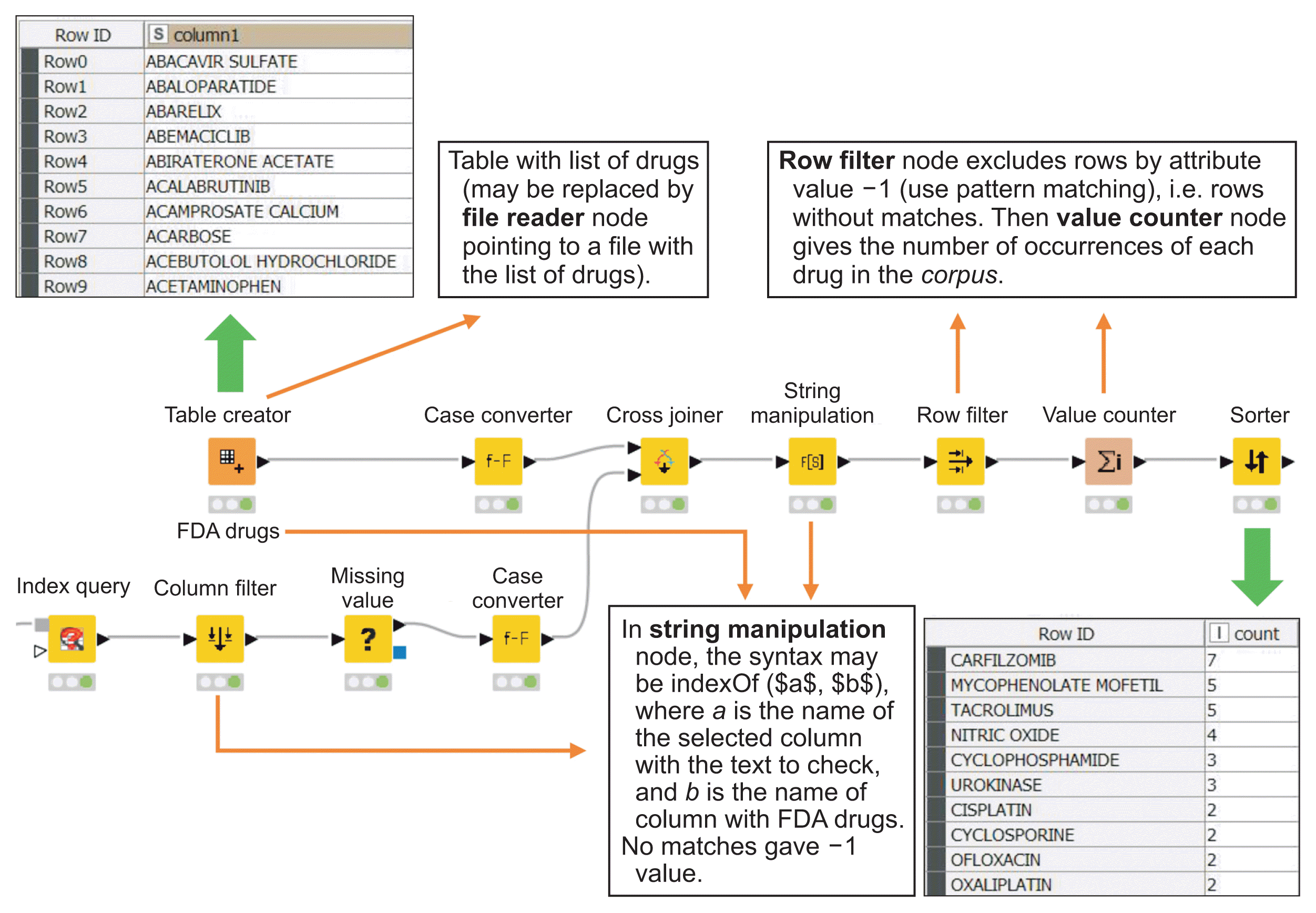
Table 1
Examples of syntax of query text in the KNIME Index Query node
The logical operators AND, AND NOT, OR, OR NOT can be used in the query that is based on Apache Lucene (https://lucene.apache.org/).
Table 2
Description of the text mining nodes used in the tutorials (based on the KNIME node descriptions)




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download