INTRODUCTION
MACHINE LEARNING
DEEP LEARNING AND CONVOLUTIONAL NEURAL NETWORK
ARTIFICIAL INTELLIGENCE IN GASTROINTESTINAL ENDOSCOPY
Table 1.
| Type | Aims | Dataset | Result | Study |
|---|---|---|---|---|
| Colonoscopy | Polyp detection | 14,000 Training images from 100 colonoscopy videos | Sensitivity, 98.79%; specificity, 98.52%; accuracy, 98.65% | Billah et al. (2017)13 |
| Polyp detection | 1,104 Nonpolyp images, 826 polyp images including white-light and narrow-band imaging | Accuracy, 87.3% | Zhang et al. (2017)14 | |
| Polyp detection (real-time) | 223 Narrow-band videos for training, 40 videos for validation, 125 videos for testing | Sensitivity, 98%; specificity, 83%; accuracy, 94% | Byrne et al. (2019)15 | |
| NICE classification | ||||
| Real-time computer-aided detection of adenoma | Randomized clinical trial | 30% Increase of adenoma detection | Repici et al. (2020)16 | |
| Differentiating adenomas from hyperplastic polyps | Separate series of 125 videos of consecutively encountering diminutive polyps | Accuracy, 94%; sensitivity, 98%; specificity, 83%; NPV, 97%; PPV, 90% | Komeda et al. (2021)17 | |
| Polyp detection | 73 Colonoscopy videos (total duration, 997 min; 1.8 million frames) from 73 patients, which included 155 colorectal polyps | Accuracy, 76.5%; sensitivity, 90.0%; specificity, 63.3% | Misawa et al. (2018)18 | |
| Polyp detection | Training data from 1,290 patients, and validation data from 27,113 images from 1,138 patients | Sensitivity, 94.38%; specificity, 95.92%; AUROC, 0.984 | Wang et al. (2018)19 | |
| EGD | Helicobacter pylori infection | 596 From 74 patients negative for infection and 65 patients positive | Sensitivity, 86.7%; specificity, 86.7%; AUROC, 0.96 | Itoh et al. (2018)20 |
| Helicobacter pylori infection | 32,208 From 1,015 patients negative for infection and 753 patients positive | Sensitivity, 88.9%; specificity, 87.4%; accuracy, 87.7% | Shichijo et al. (2017)21 | |
| Endocytoscopy | 69,142 Images (520-fold magnification) for training | Sensitivity, 96.9%; specificity, 100.0%; accuracy, 98% | Kudo et al. (2020)22 | |
| Detection of early neoplastic lesions in Barrett’s esophagus | 100 Images from 44 patients with Barrett’s esophagus | Sensitivity, 0.86; specificity 0.87 | van der Sommen et al. (2016)23 | |
| Detection of esophageal cancer | 8,428 Training images of esophageal cancer from 384 patients, 1,118 test images for 47 patients with 49 esophageal cancers and 50 patients without esophageal cancer | Sensitivity, 98%; PPV, 40%; NPV, 95%; accuracy, 98% | Horie et al. (2019)24 | |
| Identifying and delineating EGC | Retrospectively collected and randomly selected 66 EGC M-NBI images and 60 non-cancer M-NBI images into a training set and 61 EGC M-NBI images and 20 non-cancer M-NBI images into a test set | Accuracy, 96.3%; PPV, 98.3%; sensitivity, 96.7%; specificity, 95% | Kanesaka et al. (2018)25 | |
| Detecting EGC without blind spots during EGD | 3,170 Gastric cancer and 5,981 benign images to train the DCNN to detect EGC | Accuracy, 92.5%; sensitivity, 94.0%; specificity, 91.0%; PPV, 91.3%; NPV, 93.8% | Wu et al. (2019)26 | |
| 24,549 images from different parts of stomach to train the DCNN to monitor blind spots | ||||
| Determining EGC invasion depth and screening patients for endoscopic resection | 790 Images for a development dataset and another 203 images for a test dataset | AUROC, 0.94; sensitivity, 76.47%; specificity, 95.56%; accuracy, 89.16%; PPV, 89.66%; NPV, 88.97% | Zhu et al. (2019)27 | |
| Capsule endoscopy | Multiple lesion detection | 158,235 Images for training | Sensitivity, 99.8%; specificity, 100.0% | Ding et al. (2019)28 |
| 113,268,334 Images for testing | ||||
| Small-bowel hemorrhage | 9,672 Images for training | Sensitivitiy, 98.9%; recall (true positive rate) 100.0% | Li et al. (2017)29 | |
| 2,418 Images for testing | ||||
| Small-bowel erosions | 5,360 Images for training | Sensitivity, 88.2%; specificity, 90.9%; accuracy, 90.8% | Aoki et al. (2019)30 | |
| 10,440 Images for testing | ||||
| Crohn’s disease | 14,112 Images for training | Sensitivity, 96.8%; specificity, 96.6%; accuracy, 96.7% | Klang et al. (2020)31 | |
| 3,528 Images for testing |
NICE, Narrow-band Imaging International Colorectal Endoscopic; NPV, negative predictive value; PPV, positive predictive value; AUROC, area under the receiver operating characteristic curve; EGD, esophagogastroduodenoscopy; EGC, early gastric cancer; M-NBI, magnifying endoscopy with narrow-band imaging; DCNN, deep convolutional neural network.
CONSIDERATIONS FOR THE BUILDING OF AN AI-LEARNING DATASET
The establishment of separate training, validation and test sets
Sample size and class imbalance problems
The problem of overfitting
Data preparation, curation, and annotation
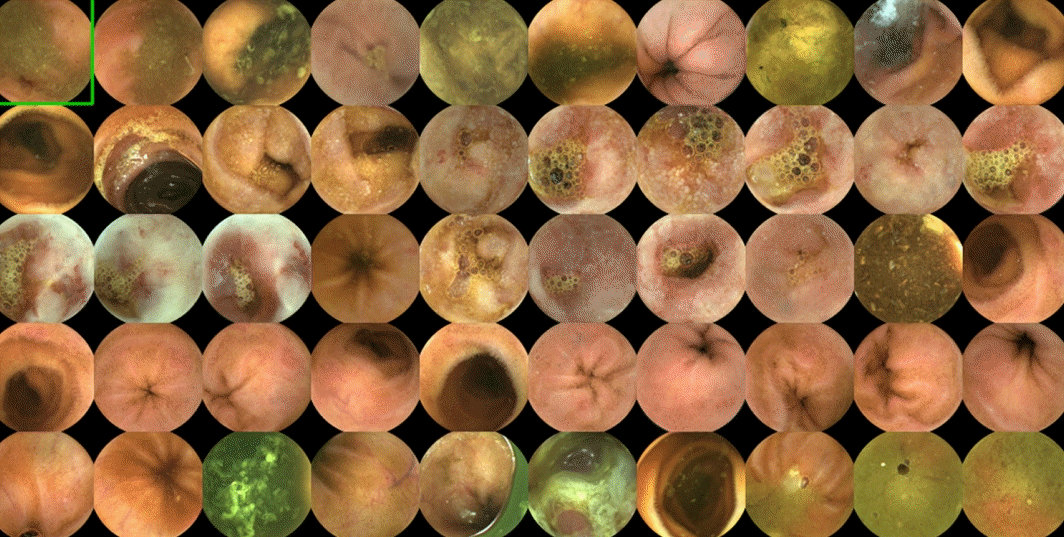 | Fig. 3.Annotation process of capsule endoscopy images for the development of convolutional neural networks. Three categorial numbers will be applied to each image in terms of medical significance, degree of protrusion and type of lesion (e.g., vascular, inflammatory, polypoid) according to a predefined reference standard. |




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download