INTRODUCTION
Carbapenem is a partially modified chemical structure of β-lactam antibiotics. It is very stable to β-lactamases and it is not affected by bacterial resistance through β-lactamases (1). In addition, it has one of the strongest antibacterial activities against gram-negative bacteria among existing antibiotics and is widely used in the medical field (2).
However, these carbapenem also developed resistant bacteria. CRE stands for Carbapenem-resistant Enterobacteriaceae, and refers to Enterobacteriaceae that have acquired resistance to Carbapenem antibiotics such as Imipenem, Meropenem, Doripenem, and Ertapenem (3).
At this point in time, CRE infection is a major medical-related infectious disease and is rapidly increasing not only in Korea but also worldwide (2). This increase in CRE infection can cause the incapacitation of the last generation antibiotic carbapenem, which was considered the last bastion of treating infectious diseases, and is a factor in the failure of microbial infection treatment in the medical field (3, 4).
The major Carbapenem-resistance mechanism of CRE is the production of Carbapenemase that is a Carbapenem- degrading enzyme, and the Enterobacteriaceae that produce Carbapenemase are called Carbapenemase-producing Enterobacteriaceae (CPE). CPE can be classified as Ambler Class according to the Carbapenemase gene amino acid homogeneity. According to this classification, the CPE genotypes are classified into Class A (Extended spectrum β-lactamases) including KPC (Klebsiella pneumoniae carbapenemase), GES (Greece extended-spectrum β-lactamase), SME (Serratia marcescens enzyme), Class B (metallo-β-lactamase) including IMP (Imipenemase), VIM (Verona integrated- encoded MBL), NDM (New Delhi MBL), SPM (Sao Paulo MBL), GIM (Germany imipenemase), SIM (Seoul imipenemase), and Class D (Oxacillinase) including OXA (Oxacillinase), and it has been reported that the distribution of major CPE genotypes varies by country (5).
The classifications that are used in this study in order to facilitate the comparison of the occurrence trends of carbapenem- resistant Enterobacteriaceae through carbapenem-degrading enzyme production and Enterobacteriaceae resistant through other mechanisms are as follows. ‘CRE (Carbapenem-Resistant Enterobacteriaceae)’ refers to all Enterobacteriaceae that exhibit antibiotic resistance regardless of the type of antibiotic resistance mechanism and ‘CPE (Carbapenemase-Producing Enterobacteriaceae)’ refers to all carbapenemase-producing Enterobacteriaceae regardless of antibiotic resistance. The aforementioned ‘CP-CRE (Carbapenemase-Producing CRE)’ refers only to the Enterobacteriaceae that are resistant to antibiotics through the mechanism of carbapenem-degrading enzyme production. In addition, ‘non CP-CRE (Non Carbapenemase-Producing CRE)’ classification to indicate the Enterobacteriaceae that exhibits antibiotic resistance through a mechanism other than the production of carbapenemase and ‘non CR-CPE (Non Carbapenem-Resistant CPE) classification to indicate the Enterobacteriaceae that has a gene encoding carbapenemase, but antibiotic resistance has not yet been expressed are used.
And the increase in Carbapenemase-producing CRE (CP-CRE) mentioned above, which has Carbapenem resistance through the production of Carbapenemase, is mainly achieved through the transmission of the CPE gene embedded in the plasmid, and the horizontal transmission of the CPE gene, which is active between Gram-negative bacteria as one of the transfers of genetic material, is known to be the direct cause of inter-microbial transmission, human-to-human transmission, and transmission within medical institutions (6).
To control and investigate CRE propagation, Korea Disease Control and Prevention Agency classified CRE infections as medical-related infectious diseases in 2011, designated them as third-tier legal infectious diseases since June 2017, and has been monitoring them thoroughly since January 2020 by reorganizing them into second-tier infectious diseases that need to be reported and quarantined within 24 hours (7). Accordingly, Ulsan Metropolitan City has also been participating in the full monitoring of CRE infections since 2018. Besides, since it had become necessary to investigate the characteristics of CRE infections in Ulsan to manage outbreaks in the region, in this study, the results of the CRE infection test accumulated from 2018 to 2021 were analyzed to determine the characteristics of local CRE incidence, such as a tendency to develop CRE infection, resistance rate to Carbapenem antibiotics, and distribution of CRE genus and CPE gene. The tests followed the experimental method of the Korea Disease Control and Prevention Agency’s SOP on CRE infection test. And VITEK2 (BioMérieux) was used for the biological identification of bacteria, and the antibiotic resistance test followed the Clinical and Laboratory Standards Institute (CLSI) guideline (8).
Go to : 
MATERIALS AND METHODS
Analysis results and classification
From 2018 to 2021, the test results of a total of 1,272 samples commissioned for the CRE infection test to the Ulsan Metropolitan Government Research Institute of Public Health and Environment were analyzed.
CRE infection test methods
The tests are carried out according to the CRE infection test method of Korea Disease Control and Prevention Agency (9). Carbapenem-resistance was confirmed through Minimal inhibitory concentration (MIC) test with Sensititre KORN plate (Thermo Fisher Scientific, U.S.).
Bacterial identification was conducted through the use of VITEK2 (BioMérieux) and 16s rRNA gene sequence analysis. For 16s rRNA PCR, the universal 27F-1492R primers for the species-level identification were used.
The CPE genes were identified through polymerase chain reaction (PCR) using Quick Taq HS DyeMix (TOYOBO, Japan). Primers for CPE gene identification tests are shown in Table 1. Primers for CPE gene analysis were based on NCBI information and the CRE infection test method of Korea Disease Control and Prevention Agency. All the sequencings were performed by Genotech Co. (Daejeon, Korea) and Cosmogenetech Co. (Daejeon, Korea).
Table 1.
Primers used for PCR detection of carbapenemase encoding genes
Go to : 
RESULTS
Tendency of CRE incidence in Ulsan
In order to find out how the trend of CRE occurrence in Ulsan changed during the investigation period, the analysis results of the CRE infection test of the samples from 2018 to 2021 were summarized according to the classifications mentioned in the introduction and shown in the Table 2. The positive rate of ‘CRE (including CP-CRE)’ according to the Minimal inhibitory concentration (MIC) test method, which is the standard for reporting positive CRE in Korea (7), was 93.08%, and the positive rate of ‘CRE or CPE’ according to the U.S. standard (4), which satisfies the criteria for reporting positive CRE infection only with CPE gene, was 95.20%. Both ‘CRE (including CP-CRE)’ and ‘CRE or CPE’ showed a tendency to increase, but the increase rate of ‘CRE or CPE’ was slightly higher.
Table 2.
Incidence of Carbapenem resistant Enterobacteriaceae infections in Ulsan by year 2018-2021
| Year Classification | 2018 | 2019 | 2020 | 2021 |
|---|---|---|---|---|
|
No. CREa or CPEb (including CP-CRE) |
220 (92.83%) |
352 (94.37%) |
311 (95.40%) |
328 (97.62%) |
| No. CP-CREc |
140 (59.07%) |
262 (70.24%) |
216 (66.26%) |
230 (68.45%) |
|
No. CRE (including CP-CRE) |
218 (91.98%) |
345 (92.49%) |
306 (93.87%) |
315 (93.75%) |
|
No. CPE (including CP-CRE) |
142 (59.92%) |
269 (72.12%) |
221 (67.79%) |
243 (72.32%) |
| No. non CP-CREd |
78 (32.91%) |
83 (22.25%) |
90 (27.61%) |
85 (25.30%) |
| No. non CR-CPEe |
2 (0.84%) |
7 (1.88%) |
5 (1.53%) |
13 (3.87%) |
| No. negative |
17 (7.17%) |
21 (5.63%) |
15 (4.60%) |
8 (2.38%) |
| Total No. tested |
237 (100%) |
373 (100%) |
326 (100%) |
336 (100%) |
‘Non CP-CRE’, which is resistant to antibiotics due to mechanisms other than Carbapenem degrading enzyme production, decreased from 32.91% in 2018 to 25.30% in 2021. During the same period, the incidence of ‘CP-CRE’ increased from 59.07% in 2018 to 68.45% in 2021, and the incidence of ‘non CR-CPE’ increased from 0.84% to 3.87%, showing a contrast to the decrease in ‘non CP-CRE’.
Composition of ‘CRE or CPE’ classification in Ulsan
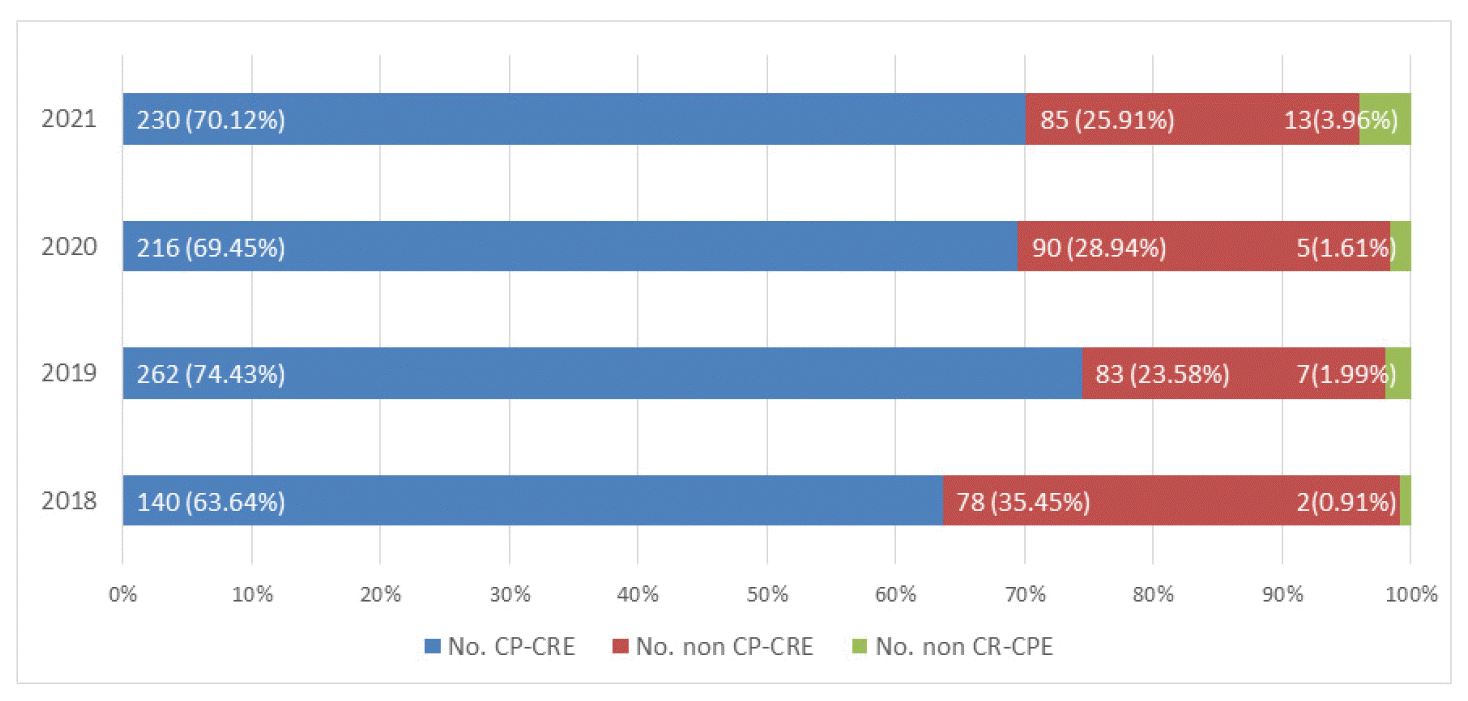
To see more closely the tendency of changes in the composition of bacteria that are Carbapenem resistant or have Carbapenem degrading enzyme genes, the percentage of ‘CP-CRE’, ‘non CP-CRE’, and ‘non CR-CPE’ are described in Fig. 1.
The ‘CP-CRE’ increased from 63.64% in 2018 to 70.12% in 2021. Over the same period, the proportion of ‘non CR-CPE’ increased from 0.91% to 3.96%, while the proportion of ‘non CP-CRE’ decreased from 35.45% to 25.91%.
Carbapenem Antibiotic resistance patterns of Carbapenem-resistant Enterobacteriaceae
Table 3 shows the analysis of resistance patterns for four types of carbapenems from 2018 to 2021. A total of 1,184 CREs were confirmed during the survey period, of which 1,117 (94.34%) showed the highest resistance rate due to Ertapenem resistance. Second, Imipenem resistance was found to be 774 cases (65.37%), followed by Meropenem resistance 750 cases (63.34%), and Doripenem resistance 624 cases (52.70%). There were 572 cases (48.31%) of CRE that showed resistance to all four Carbapenem.
Table 3.
Antibiotic susceptibility pattern of Carbapenem-resistant Enterobacteriaceae
Distribution of CRE genus in Ulsan
Distribution of CRE genus in Ulsan were investigated. Table 4 shows the distribution of CRE genus by year from 2018 to 2021 and the average distribution of CRE genus during the survey period. Klebsiella was the most separated with an average of 72.04%, followed by Escherichia with an average of 10.81%, Enterobacter with an average of 9.54% and Citrobacter with 1.27%. In addition to this, Providencia, Serratia, Morganella, Raoultella, Pantoea, Proteus, and Leiiotia were classified, and ‘No classification’ was samples that had not obtained data on CRE genus because only CPE gene tests were requested after other institutions tested positive for CRE.
Table 4.
Bacterial genus composition of the Carbapenem-resistant Enterobacteriaceae per year in Ulsan by year 2018-2021
Distribution of CPE genes in Ulsan
Following the distribution of the CRE genus, the most isolated CPE genes were also identified and shown in Table 5. Distribution of CPE genes and CPE subtypes separated from 2018 to 2021 were analysed in this table. Of the 879 CPE genes separated, the KPC gene was the most isolated with 785 (89.31%), followed by 75 NDM genes (8.53%), both OXA and IMP genes with 9 (1.02%) and 1 VIM gene (0.11%). By KPC, NDM, and OXA subtypes, the KPC-2, NDM-1, and OXA-48 ratios were the highest in order. During the investigation period (2018-2021), the average isolation rate of KPC-2 was the highest at 88.74%, the second highest in NDM-1 with 6.03%, and the third highest at NDM-5 with 2.39%.
Table 5.
Prevalence of Carbapenemase genes in Carbapenem-resistant Enterobacteriaceae from 2018-2021
|
Classification (Total 879) |
Subclassification | 2018 | 2019 | 2020 | 2021 | sum |
|---|---|---|---|---|---|---|
|
KPC 785 (89.31%) |
KPC-2 |
131 a, b (90.97%) |
257 (95.19%) |
202 (91.40%) |
190 d (77.87%) |
780 |
| KPC-3 | 1 (0.69%) | - | - |
1 (0.41%) |
2 | |
| KPC-4 | - | - | - |
3 (1.23%) |
3 | |
|
NDM 75 (8.53%) |
NDM-1 |
3 a (2.08%) |
2 (0.74%) |
7 (3.17%) |
41 (16.8%) |
53 |
| NDM-4 | - | - | 1 (0.45%) | - | 1 | |
| NDM-5 |
7 (4.86%) |
6 c (2.22%) |
5 (2.26%) |
3 (1.23%) |
21 | |
|
OXA 9 (1.02%) |
OXA-48 | 2 b (1.39%) |
2 (0.74%) |
1 (0.45%) |
- | 5 |
| OXA-181 | - |
2 c (0.74%) |
1 (0.45%) |
- | 3 | |
| OXA-232 | - | - |
1 d (0.41%) |
1 | ||
|
IMP 9 (1.02%) |
IMP-4 | - |
1 (0.37%) |
3 (1.36%) |
5 (2.05%) |
9 |
|
VIM 1 (0.11%) |
VIM-2 | - | - |
1 (0.45%) |
- | 1 |
|
No. CPE positive (including CP-CRE) |
142 | 269 | 221 | 243 | - | |
In addition to this, two ‘Klebsiella pneumoniae’ in 2018, one with both KPC-2 and NDM-1 CPE and the other with both KPC-2 and OXA-48, one ‘Escherichia coli’ in 2019 with both NDM-5 and OXA-181, and one ‘Citrobacter fruendii’ in 2021 with both KPC-2 and OXA-232 were identified.
Distribution of CPE genes according to genera and species of CRE
Table 6 shows the distribution of CPE genes by CRE genus and species. By 2020, there were four separate genus of bacteria: Klebsiella, Escherichia, Enterobacter, and Citrobacter, but in 2021, they were further separated from three genus: Raoultella, Morganella, and Proteus.
Table 6.
Distribution of Carbapenemase genes according to genera of Carbapenem-resistant Enterobacteriaceae from 2018 to 2021
| Year | CPE Genus | KPC | NDM | OXA | IMP | VIM |
|---|---|---|---|---|---|---|
| 2018 |
Klebsiella pneumoniae (109) |
109 KPC-2(108)a, b KPC-3(1) |
1 NDM-1(1)a |
1 OXA-48(1)b |
- | - |
|
Escherichia coli (15) |
8 KPC-2(8) |
6 NDM-1(1), NDM-5(5) |
1 OXA-48(1) |
- | - | |
|
Enterobacter spp. (4) |
3 KPC-2(3) |
1 NDM-1(1) | - | - | - | |
|
Enterobacter cloacae (4) |
4 KPC-2(4) |
- | - | - | - | |
|
Enterobacter aerogenes (1) |
1 KPC-2(1) |
- | - | - | - | |
|
Citrobacter freundii (2) |
1 KPC-2(1) |
1 NDM-5(1) |
- | - | - | |
|
Citrobacter amalonaticus (1) |
- |
1 NDM-5(1) |
- | - | - | |
| 2019 |
Klebsiella pneumoniae (217) |
216 KPC-2(216) |
- |
1 OXA-48(1) |
- | - |
|
Escherichia coli (36) |
26 KPC-2(26) |
8 NDM-1(2), NDM-5(6)c |
3 OXA-48(1)c, OXA-181(2) |
- | - | |
|
Enterobacter cloacae (11) |
10 KPC-2(10) |
- | - |
1 IMP-4(1) |
- | |
|
Enterobacter aerogenes (2) |
2 KPC-2(2) |
- | - | - | - | |
|
Citrobacter freundii (1) |
1 KPC-2(1) |
- | - | - | - | |
|
Citrobacter youngae (1) |
1 KPC-2(1) |
- | - | - | - | |
|
Citrobacter farmeri (1) |
1 KPC-2(1) |
- | - | - | - | |
| 2020 |
Klebsiella pneumoniae (155) |
150 KPC-2(150) |
5 NDM-1(4), NDM-5(1) |
- | - | - |
|
Klebsiella ozaenae (1) |
1 KPC-2(1) |
- | - | - | - | |
|
Escherichia coli (20) |
13 KPC-2(13) |
5 NDM-1(1), NDM-4(1), NDM-5(3) |
2 OXA-148(1), OXA-181(1) |
- | - | |
|
Enterobacter cloacae (11) |
6 KPC-2(6) |
1 NDM-1(1) |
- |
3 IMP-4(3) |
1 VIM-2(1) |
|
|
Citrobacter freundii (2) |
1 KPC-2(1) |
1 NDM-1(1) |
- | - | - | |
|
Citrobacter amalonaticus (1) |
1 KPC-2(1) |
- | - | - | - | |
|
Citrobacter braakii (1) |
1 KPC-2(1) |
- | - | - | - | |
| 2021 |
Klebsiella pneumoniae (175) |
150 KPC-2(147)e, KPC-3(1), KPC-4(2) |
25 NDM-1(25) |
- | - | - |
|
Klebsiella oxytoca (4) |
- |
4 NDM-1(4) |
- | - | - | |
|
Escherichia coli (14) |
8 KPC-2(8) |
6 NDM-1(3), NDM-5(3) |
- | - | - | |
|
Enterobacter cloacae (13) |
3 KPC-2(3) |
5 NDM-1(5) |
- |
5 IMP-4(5) |
- | |
|
Enterobacter aerogenes (3) |
2 KPC-2(2) |
1 NDM-1(1) |
- | - | - | |
|
Enterobacter asburiae (1) |
1 KPC-2(1) |
- | - | - | - | |
|
Citrobacter freundii (6) |
5 KPC-2(4)d, KPC-4(1) |
1 NDM-1(1) |
1 OXA-232(1)d |
- | - | |
|
Raoultella planticola (1) |
- |
1 NDM-1(1) |
- | - | - | |
|
Morganella (1) |
1 KPC-2(1) |
- | - | - | - | |
|
Proteus (1) |
- |
1 NDM-1(1) |
- | - | - |
In addition, CPE genes tended to vary. In 2018, KPC was the most isolated in Klebsiella, and all but one KPC-3 was identified as KPC-2. NDM-1 and OXA-48 were isolated one each, and a total of four CPE genes were isolated from the genus Klebsiella. And in 2021, two KPC-4 were first separated from Klebsiella, and the number of NDM-1 separations increased from 1 in 2018 to 29.
In Escherichia, various KPC, NDM, and OXA genes were isolated, and NDM-4 was isolated for the first time in 2020. In 2021, NDM-1 was isolated in new genus, such as Raoultella and Proteus. For Citrobacter, it showed relatively diverse CPE gene isolates despite low CRE resistance confirmation rates or CPE gene isolates.
Furthermore, in 2018 and 2019, CPE genes were separated from the single Klebsiella pneumoniae species, but in 2020, CPE genes were also detected in Klebsiella ozaenae and oxytoca species. Escherichia coli was found to be the most diverse kinds of CPE genes isolated, such as KPC-2, NDM-1, NDM-4, NDM-5, OXA-28, and OXA-181, and IMP-4 and VIM-2 classified as Ambler class B were isolated only in Enterobacter cloacae.
Go to : 
DISCUSSION
Enterobacteriaceae are bacteria that also inhabit the intestines of humans and are widely distributed in medical institutions and local communities, thus Enterobacteriaceae can easily be the source of hospital-acquired and community-acquired infections. Therefore, if the number of Enterobacteriaceae that have acquired antibiotic resistance increases, it may lead to an increase in human CRE infection and pose a threat to public health (10, 11).
For this reason, in response to the need to confirm the trend of CRE in Ulsan, which can be a potential threat to local public health, this investigation was conducted. And the study confirms that the incidence of CRE is increasing in Ulsan. In addition, as we can see in Table 5 and 6, it was confirmed that the increase in CRE in Ulsan was driven by the increase in CPE. Although there are several mechanisms of Carbapenem resistance, increased resistance by the Carbapenemase enzyme is much more difficult to control transmission than other mechanisms of Carbapenem resistance.
Mechanisms of antibiotic resistance in bacteria include decreased outer membrane permeability, overexpression of efflux pumps, changes in the receptor structure on which antibiotics act, self-destruction of antimicrobial substances, or a combination of the aforementioned mechanisms (12). The mechanisms by which Enterobacteriaceae resist Carbapenem antibiotics are also the same. In particular, the production of Carbapenem-degrading enzymes is pointed out as the most important Carbapenem antibiotic resistance mechanism, and resistance propagation by this mechanism is rapidly taking place worldwide (13, 14). Because most CPE genes are embedded in mobile genetic structures such as insertion sequences, integrons, and transposons that can spread horizontally between gram-negative bacteria, so they are easy to spread to another (15). Through the horizontal gene transfer, Gram-negative bacteria increases the incidence of CP-CRE infection in medical facilities worldwide (16). In addition, there are many studies on the risks of CP-CRE, including a paper finding that CP-CRE bacteremia patients have a significantly higher mortality rate than non-CP-CRE patients (17), so it is necessary to prepare countermeasures against the increase in the incidence of CP-CRE.
However, the increase in CPE shown in this study supports that active CPE gene transfer is taking place not only in many other cities and countries but also in Ulsan, and it is highly expected that the acquisition of antibiotic resistance by CPE gene transfer will continue to increase in the future.
Next, the tendency (Ertapenem-Imipenem-Meropenem-Doripenm) of the samples in Ulsan to be resistant to the four types of Carbapenem was confirmed to be the same as the national tendency announced by the Korea Centers for Disease Control and Prevention (18). It was conspicuous that the resistance to Ertapenem was high enough to account for 94.43% of the total antibiotic resistance. In addition, it was investigated that there was no bacteria that was susceptible to Ertapenem and showed resistance to all three of Imipenem, Meropenem, and Doripenem at the same time from the samples in Ulsan.
The following is a review of the CRE genus and species. The most frequently confirmed bacteria as CRE in Ulsan was Klebsiella pneumoniae, which was also confirmed to be the same as the nation’s largest CRE bacteria announced by the Korea Centers for Disease Control and Prevention (18). Klebsiella pneumoniae is a genus of Enterobacteriaceae that has played a major role in the transmission of extended-spectrum b-lactamases (ESBLs) genes in healthcare institutions for the past 30 years. Based on this, it is expected to contribute a lot to the propagation of Carbapenemase (11). In addition, several studies have shown that patients infected with carbapenem-resistant Klebsiella pneumoniae is more likely to die than patients infected with carbapenem-sensitive Klebsiella pneumonia (19, 20).
Further more, identifying not only the type of CRE bacteria but also the type of carbapenemase produced by CRE bacteria is also the focus of CRE management.
KPC is the most widely distributed type of CPE not only in Korea but also worldwide. KPC is found in many Enterobacteriaceae such as Escherichia, Proteus, Serratia, Salmonell, and Citrobacter (21), and KPC was isolated the most in this study as well. The bacteria that produce KPC are occasionally found in local communities, but they are a representative medical-related infection source that is often spread during health care such as antibiotic treatment, long-term hospitalization, or organ transplant surgery (22, 23, 24).
Among the KPC subtypes, KPC-2 and KPC-3 appear most frequently, and KPC-2 is the most prevalent worldwide (21). In Asia, the KPE gene is indigenous to Korea and China, and it is reported that KPC is found in various Gram-negative bacteria in those countries (25, 26). It is reported that the mortality rate due to infection with these KPC-producing bacteria is over 50% (27, 28, 29).
In the case of NDM-1, it increased from 3 cases in 2018 to 41 cases in 2021, leading to an increase in the overall NDM separation rate in this study. Accordingly, the isolation rate of KPC-2, which exceeded 90% from 2018 to 2020, showed a relatively decreasing trend to 77.87% in 2021.
NDM-1 is a metallo-β-lactamase belonging to Ambler class B, which existed endemic to the Indian subcontinent in the past, but has spread rapidly and has recently been reported around the world. Unlike other β-lactamases having a Serine group, this is a very stable structure combined with Zinc, and is known as a very strong antibiotic resistance gene capable of degrading almost all β-lactam antibiotics including Carbapenem (30, 31). A study result was also announced that the plasmid (Inc A/C type) delivering the NDM-1 gene can contain up to 14 other antibiotic resistance factors together (32, 33, 34). Furthermore, this gene can be spread not only to intestinal bacteria, but also to various bacteria that exist in the natural environment, so there is a concern that it will continue to spread strong antibiotic resistance rapidly (34, 35, 36).
In addition, bacteria with two or more CPE genes were found through CPE gene distribution investigations by CRE genus and species.
Bacteria having several antibiotic resistance factors, including Carbapenem degrading enzyme, have been steadily reported worldwide, and bacteria having two or more CPE genes have already been reported several times in Korea (37, 38, 39). Since not all Carbapenemase can degrade all β -lactam antibiotics, it can be treated with limited but still sensitive antibiotics, However, the antibiotic resistance range of bacteria that can produce two or more Carbapenem-degrading enzymes is widened, and there are not many available antibiotics left, which can become a big difficulty in patient treatment (37).
In conclusion, through the CPE gene distribution investigations by CRE genus and species, it was also confirmed that the CRE genus and species in Ulsan were diversified. And it was found that not only the CRE species from which the CPE gene was isolated, but also the CPE gene subtypes separated from CRE were diversified. Unfortunately, this diversification is being observed worldwide, making it more difficult to select an appropriate antibiotic and dose (31).
As this study shows, as the increase in the incidence of CRE and the spread of CPE genes are rapidly increasing and becoming intractable everywhere, many efforts are urgently needed to prevent the spread of CRE, a threat to public health. Antibiotic doses should be minimized by activating stewardship on the use of antibiotics and prevention and management of the spread of infectious diseases in public health should be carried out thoroughly (40). Early detection of CRE through accurate laboratory diagnosis is also important (10). In addition, due to the biological nature of bacteria, not only the medical field but also cross-ministerial efforts of the One-health concept must be accompanied (41).
Through this investigation, the characteristics of CRE infection in Ulsan were confirmed, and it is expected that it can be used as basic data for preparing appropriate CRE infection management measures suitable for the situation of the local community and prescribing empirical antibiotics.
Go to : 




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download