INTRODUCTION
The etiological agent of porcine epidemic diarrhea (PED) is the porcine epidemic diarrhea virus (PEDV). It is a large, enveloped RNA virus and is a member of the genus Alphacoronavirus belonging to the Coronaviridae family placed with the Arteriviridae family in the order Nidovirales on the basis of similarities in genome organization and replication strategy (1, 2). PEDV has a positive single-stranded RNA molecule of approximately 30 kb in length (2).
PEDV encodes two nonstructural polyproteins (pp1a and pp1ab), four structural proteins (the approximately 200-kDa glycosylated spike (S) protein, 8-kDa envelope (E) protein, 24-kDa membrane (M) protein, and 58-kDa nucleocapsid (N) protein), and one known accessory protein (the open reading frame (ORF) 3 protein) (3).
Coronavirus E protein forms cation-selective ion channels that are more permeable to monovalent cations than to monovalent anions. It plays a significant role in viral particle budding. On the other hand, N protein interacts with viral genomic RNA and then forms the helical nucleocapsid during viral particle assembly. M protein is required in the assembly process and can elicit the production of antibodies. The accessory protein encoded by ORF3 is thought to serve as an ion channel and can affect virus production and virulence. S protein is the major envelope type I glycoprotein of virion (4, 5, 6).
Swine enteritis in all ages is caused by PEDV, transmissible gastroenteritis virus (TGE), rotavirus, Eimeria spp. etc., and is often fatal among neonatal piglets. PEDV infection causes diarrhea, resulting in up to 100% mortality in piglets (7). The histologic lesions of PEDV are characterized by villus atrophy and fusion in the jejunum and ileum (8).
In Korea, the increase in the incidence of diarrhea and death in neonatal piglets associated with PEDV infection has become a major economic concern.
The intestine comprises an epithelial monolayer, which acts as a barrier between the outside, including the microbiota, and the host (9). The epithelial barrier plays fundamental roles in immune defenses against enteric viral infections to prevent disease by integrating diverse signals, including those from the microbiota (9). Electron microscopy was employed in some of the earliest reports of enteric viruses targeting goblet cells and neighboring epithelial cells in the gastrointestinal system (10). PED has caused tremendous losses in the pig industry in the United States; outbreaks were also reported in Central Europe (11).
Studies comparing wt-PEDV and ca-PEDV strains have not yet been conducted. Thus, this study aimed to compare the pathogenicity and nucleotide sequence of ORF3 between wt-PEDV and ca-PEDV.
Go to : 
MATERIALS AND METHODS
PEDV strains
In experimental pig inoculation, the wt-PEDV strain SNUVR971496 and ca-PEDV derived from that strain were used (12). A 3-day-old piglet with severe diarrhea was used to perform isolation of wt-PEDV from the small intestines of the piglet. Serial 10-fold dilutions in Vero cells were used to titrate wt-PEDV by growth, followed by viral antigen with PEDV monoclonal antibody immunofluorescence labeling. The monoclonal antibody reacted with the 30-kDa membrane protein of PEDV in western blot analysis (13). PEDV adaptation was confirmed in Vero cells (12, 14).
Experimental design
We randomly divided 45 1-day-old colostrum-deprived pigs into three groups.
A total of 15 pigs from each treatment group and 15 pigs from the negative control group were orally inoculated with a 5-mL disposable syringe with 2 mL of virus stock containing 1 × 106 tissue culture infective doses of wt-PEDV and ca-PEDV, respectively. About 15 negative control pigs were orally administered with 2-mL uninfected cell culture supernatant fluid. The piglets were placed in stainless steel isolator (3 piglets each isolator), sterilized with accelerated hydrogen peroxide (McKesson Medical–Surgical Inc., Richmond, USA) disinfectant, and fed a sterile milk substitution (SPF-LacⓇ, Borden, Hampshire, IL, USA). All piglets were examined for clinical signs of diarrhea, dehydration, anorexia, and vomiting three times daily. From the infected and control groups, three piglet subgroups were killed by electrocution and subjected to necropsy at 12, 24, 36, 48, and 60 h post inoculation (hpi) (12). The Institutional Animal Care and Use Committee (SNU-210203-1) collected the tissue specimens at a facility in Seoul National University, Suwon.
Virus isolation
Wt-PEDV and ca-PEDV were isolated from small intestinal tissues (6, 8). Scraped-off mucosa and small intestine contents were pooled, diluted 1:5 in phosphate-buffered saline (PBS; 0.15 M NaCl, 0.01 M phosphate buffer [pH 7.4]), and homogenized via ultrasonication. The result contained PEDV suspension clarified via centrifugation at 10,000 × g for 5 min to prevent cell culture bacterial or fungal contamination (15).
In situ hybridization
A 412 base pair cDNA probe was used to encode viral RNA of the PEDV membrane protein. The forward and reverse primers were 5'-GGGCGCCTGTATAGAGTTTA-3' (nucleotides 927 to 946) and 5'-AGACCACCAAGAATGTGTCC-3' (nucleotides 1319 to 1338), respectively. Reverse transcription polymerase chain reaction (RT-PCR) was conducted (13, 16). The Wizard PCR Preps Purification System was used (Promega Biotech, Madison, WI, USA) to purify RT-PCR products and labeled via digoxigenin–dUTP random priming using a commercial kit (Boehringer Mannheim, Indianapolis, IN, USA) according to the manufacturer’s protocols. In situ hybridization was performed (17). The digoxigenin-labeled transmissible gastroenteritis virus (TGEV) cDNA probe was used as a negative probe (18). Negative tissue (jejunum and ileum) controls were collected from three 1-day-old colostrum-deprived piglets experimentally infected with TGEV (17).
Sequencing
Based on the CV777 ca-PEDV design (GenBank accession no. AF353511) by Dr. Kurt Tobler (Institute of Virology, University of Zurich, Switzerland), primers were used to amplify the whole ORF3 genome. 5'-TCCTAGACTTCAACCTTAC-3' (nucleotides 24741 to 24759) and 5'-TCATAGACCATTATCATTCA-3' (nucleotides 25476 to 25457) were the forward and reverse primers, respectively. These primers amplified the 733 base pair cDNA fragment. RNA was extracted from wt-PEDV- and ca-PEDV-infected cells, and sequencing was performed using a commercial service (BioFact, Daejeon, Korea).
Morphometric analysis
A total of three pieces of formalin-fixed jejunum were taken from each virus-infected and control piglet for morphometric analysis (17).
Statistical analysis
The Wilcoxon matched-pairs signed rank test and one-way analysis of variance (ANOVA) test were employed to determine the significance of the differences between infected and control piglets in terms of the villus height/crypt depth (VH/CD) ratio and the mean number of positive cells for ISH per villus (12). The Wilcoxon test confirmed the change between groups by the mean and standard deviation comparing before and after, not the mean and standard error (12).
Furthermore, result from ANOVA test which showed statistical significance was evaluated by conducting a post hoc test for a pairwise comparison with Tukey’s adjustment.
Go to : 
RESULTS
Clinical signs
Two piglets from the wt-PEDV-infected group and one piglet from the ca-PEDV-infected group started experiencing diarrhea at 12 hpi. Diarrhea was observed in seven piglets in wt-PEDV-infected group and six piglets in the ca-PEDV- infected group at 36 hpi; however, at 48 and 60 hpi, all piglets in the wt-PEDV- and ca-PEDV-infected groups had diarrhea. Anorexia and dehydration were observed in piglets at the onset of diarrhea in both groups. Vomiting was noted in some piglets from both groups at 36 hpi. No clinical signs were observed in the control piglets. Also, changes in weight and body temperature were not detected.
Villus height/crypt depth ratio
Mean jejunal villus height and crypt depth (VH/CD) ratio in piglets infected orally with Korean strain of PEDV were 6.8 ± 0.8, 7.0 ± 0.8, 8.2 ± 1.0, 8.0 ± 1.3, and 7.9 ± 1.1 in uninfected control piglets; 7.2 ± 1.2, 5.2 ± 1.4, 2.7 ± 0.3, 1.4 ± 0.2, and 1.5 ± 0.1 in wt-PEDV-infected piglets; and 7.6 ± 1.8, 3.2 ± 0.4, 2.2 ± 0.3, 1.3 ± 0.3, and 1.1 ± 0.2 in ca-PEDV-infected piglets at 12, 24, 36, 48, and 60 h post inoculation (Table 1).
Table 1.
Mean jejunal villus height and crypt depth (VH/CD) ratio in piglets infected orally with Korean strain of porcine epidemic diarrhea virus (PEDV)
|
Hours post-inoculation |
Mean VH/CD ratio (±SD) in each groups | ||
|---|---|---|---|
| Uninfected control piglets | Vero cell adapted -PEDV- infected piglets | Wild type-PEDV- infected piglets | |
| 12 | 6.8±0.8 | 7.2±1.2 | 7.6±1.8 |
| 24 | 7.0±0.8a* | 5.2±1.4a | 3.2±0.4b |
| 36 | 8.2±1.1a | 2.7±0.3b | 2.2±0.3b |
| 48 | 8.0±1.3a | 1.4±0.2b | 1.3±0.4b |
| 60 | 7.9±1.1a | 1.5±0.1b | 1.1±0.2c |
The mean VH/CD ratios in piglets experimentally inoculated with wt-PEDV and ca-PEDV were significantly different (P < 0.05) from those of the control piglets. Furthermore, the mean scores for wt-PEDV-infected piglets were significantly different (P < 0.05) from those of ca-PEDV-infected piglets at 24, 36, and 60 hpi.
Antigen-positive cells per villus
Statistical analysis of the mean positive scores of the jejunal tissues revealed significant differences in the amount of PEDV nucleic acid between wt-PEDV and ca-PEDV. ISH positive results in piglets orally infected with the Korean strain of PEDV jejunal villus were 4.0 ± 0.8, 10.3 ± 0.5, 7.3 ± 0.5, 7.0 ± 0.8, and 5.3 ± 0.5 in ca-PEDV-infected piglets; and 4.3 ± 0.9, 16.7 ± 1.2, 9.3 ± 0.5, 9.0 ± 0.8, and 5.7 ± 0.9 in wt-PEDV-infected piglets, respectively. These values indicate a significant difference among each group (Table 2).
Table 2.
In-situ hybridization (ISH) positive results in piglets infected orally with the Korean strain of porcine epidemic diarrhea virus (PEDV) in jejunal villus
|
Hours post-inoculation |
Numbers of positive villus cells (in subgroups of 3 piglets) by in-situ hybridization (ISH) at each hour post-inoculation | ||
|---|---|---|---|
| Uninfected control piglets | Vero cell adapted -PEDV- infected piglets | Wild type-PEDV- infected piglets | |
| 12 | 0.0±0.0a* | 4.0±0.8b | 4.3±0.9b |
| 24 | 0.0±0.0a | 10.3±0.5b | 16.7±1.2c |
| 36 | 0.0±0.0a | 7.3±0.5b | 9.3±0.5c |
| 48 | 0.0±0.0a | 7.0±0.8b | 9.0±0.8b |
| 60 | 0.0±0.0a | 5.3±0.5b | 5.7±0.9b |
A significantly higher number of PEDV-infected cells in the jejunal tissues was detected (P < 0.05) in ca-PEDV-inoculated piglets than in wt-PEDV-inoculated piglets at 24, 36, and 60 hpi.
In situ hybridization
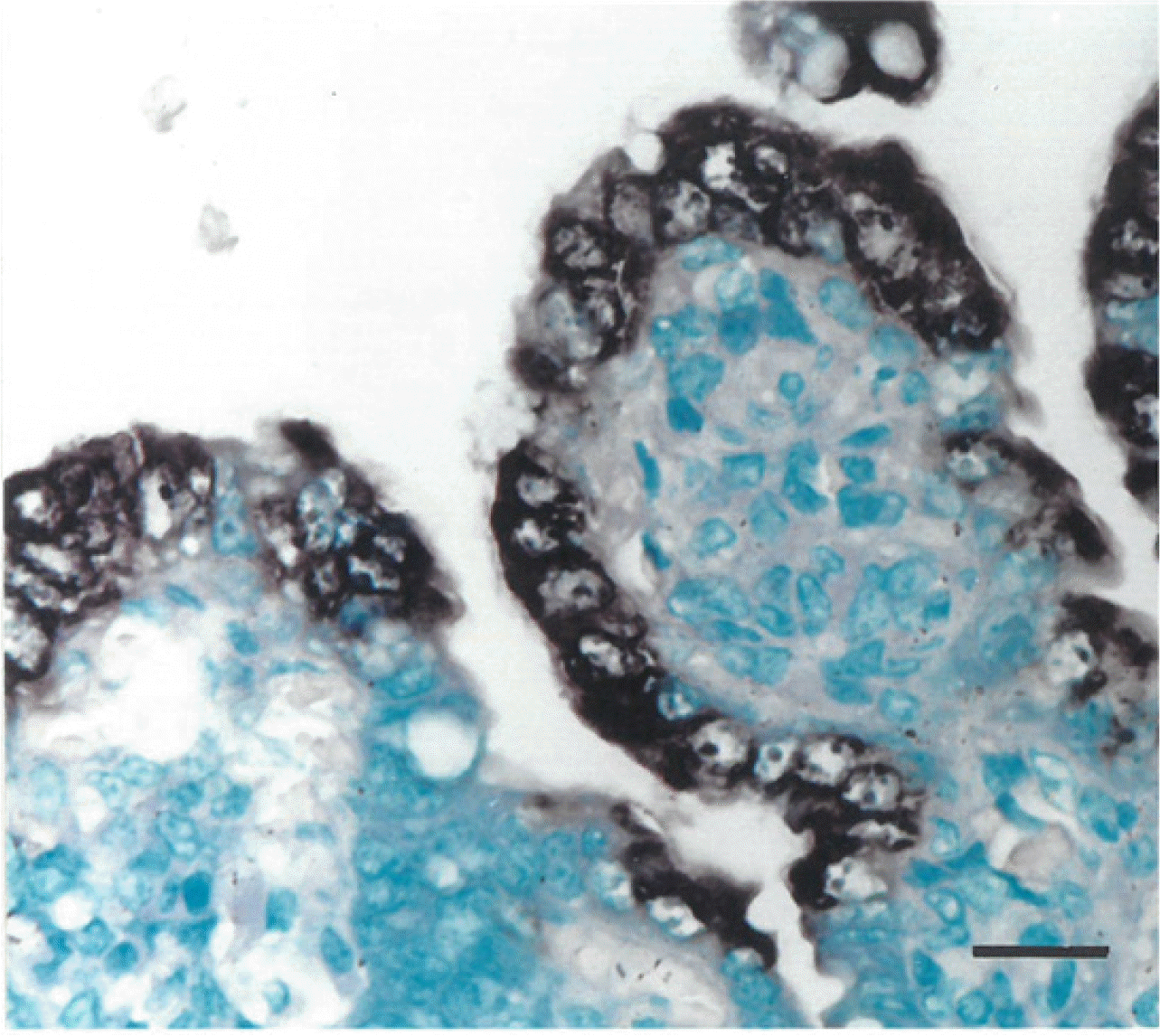
Positive cells typically exhibit a black ISH reaction product in the cytoplasm, without background staining. PEDV-positive cells were found mainly in the jejunum villus enterocytes (Fig. 1) and ileum, typically in areas of moderate-to-severe villus atrophy or vacuolation in piglets from both the wt-PEDV- and ca-PEDV-infected groups. The villi of the small intestines covered by positive cells were arranged continuously in the enterocytes. Positive cells were not detected in the epithelial cells lining the crypts of Lieberkühn. Cuboidal and degenerated or regenerated enterocytes with or without vacuolated cytoplasm were detected in some areas. Pretreatment with RNase A eliminated hybridization signal from 30 piglets experimentally infected with either wt-PEDV or ca-PEDV. No hybridization signals for PEDV were observed in sections from negative control pigs. Experimental design was carried out by quantifying the amount of virus, and the number of virus-positive cells was counted via immunostaining and ISH without measuring the titer of virus isolation.
Virus isolation
Wt-PEDV and ca-PEDV were isolated from the small intestines of piglets inoculated with PEDV at 24, 36, 48, and 60 hpi. No PEDV was isolated from piglets in the control group. Immunostaining and ISH without measurement of the titer of virus isolation was used to count the numbers of virus-positive cells: 2–4 positive villus cells at 12 hpi, 6–10 positive villus cells at 24 hpi, 12–14 positive villus cells at 36–48 hpi, and 15–16 positive villus cells at 60 hpi in the wt-PEDV group.
Nucleotide sequences
Nearly identical (98.7% homology) nucleotide sequences of wt-PEDV and ca-PEDV were noted. Nucleotide substitutions were detected in ORF3 that led to some amino acid changes (Table 3). Therefore, the coding changes observed in ca-PEDV may represent nucleotide changes reflecting the cell culture adaptation of wt-PEDV.
Table 3.
Nucleotide changes within the open reading frame 3 (ORF3) cause four differences between the predicted amino acid sequence of wild type (wt)-porcine epidemic diarrhea virus (PEDV), and cell culture adapted (ca)-porcine epidemic diarrhea virus (PEDV)
Go to : 
DISCUSSION
The results of this study indicated that wt-PEDV is significantly more virulent than ca-PEDV. The VH/CD ratio is an important marker for toxicity evaluation. Because the VH/CD ratio significantly decreased compared with that of the control group, this means that damage to the villi is an indicator of toxicity evaluation.
Wt-PEDV was found to be virulent and induced more severe clinical signs and microscopic enteric lesions than ca-PEDV. Wt-PEDV-inoculated pigs had a higher amount of viral nucleic acid in the small intestines. PEDV nucleic acid cell types were detected, and the distributions of viral nucleic acid within the small intestines were generally similar for these two viruses despite the differences in virulence. The jejunum was the most consistent and intense staining for both types of PEDV.
The wt-PEDV strain SNUVR971496 could be a virulent virus. This conclusion is consistent with the devastating disease observed on the farm from which the strain SNUVR971496 originated (12). PEDV-induced enteric disease pathogenesis is most likely caused by the destruction of villus enterocytes. Virus replication in jejunal and ileal villus enterocytes has also been described in PEDV-infected pigs (19). Epidemic PEDV strains rapidly spread and cause a high number of deaths among pigs (20).
These changes compromise the digestion and absorption functions of villus enterocytes. Fecal virus shedding (alongside frequent detection of PEDV RNA in the nasal cavity), acute viremia, severe atrophic enteritis (mainly jejunum and ileum), and increased pro-inflammatory and innate immune responses are observed in neonatal pigs with the viral infection (21).
Villus atrophy and fusion were characteristic lesions induced by PEDV in this study. The degree of morphologic changes observed in the small intestines varied depending on the time after inoculation. There was a corresponding increase in the severity of diarrhea and dehydration of the infected piglets, which began as the villus height gradually decreased. Morphometry confirmed a significant reduction in villus height in the jejunum at 60 hpi.
Both wt-PEDV- and ca-PEDV-infected enterocytes replicated within them, causing necrosis and sloughing. There were differences between the two viruses in the severity of damage to the small intestines and the amount of infection. The mean VH/CD ratios were more significantly reduced in the jejunum of wt-PEDV-inoculated piglets than in that of ca-PEDV-inoculated piglets at 36 hpi. The low rate of enterocyte loss in ca-PEDV-inoculated piglets could be the result of the inability of the virus to sufficiently infect enterocytes, or it could be because the process of virus replication did not rapidly lead to sloughing of the infected enterocytes. There is evidence that both mechanisms are contributory. The amount of PEDV nucleic acid indicated that ca-PEDV-infected fewer enterocytes and replicated slower than wt-PEDV in the early stage of infection. These results suggested that ORF3 gene alteration may cause a slower ca-PEDV replication in the enterocytes compared with wt-PEDV.
Statistically significant differences were observed in the ca-PEDV pathogenicity compared with its parental wt-PEDV. The mechanisms responsible for the different degrees of pathogenicity between wt-PEDV and ca-PEDV are not yet understood. Nucleotide changes in ORF3 were identified in ca-PEDV, which possibly influence PEDV pathogenicity. These results indicate that ORF3 may be an important determinant for in vivo virulence and in vitro replication of PEDV. The ORF3 accessory protein has been proposed as a crucial viral virulence factor in a natural host (22). Further study is needed due to the lack of an extensive comparative study of ORF3 to examine sequence differences in ORF3 between wt-PEDV and ca-PEDV.
Go to : 




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download