This article has been
cited by other articles in ScienceCentral.
Abstract
Background
Patients with hematologic malignancies may produce replication-competent virus beyond 20 days of SARS-CoV-2 infection. However, data regarding the transmission of SARS-CoV-2 from patients with prolonged viral shedding is limited.
Methods
In May 2022, four additional cases of COVID-19 were reported in a hematologic ward at a tertiary care hospital in South Korea, after an 8-week isolation of a patient with prolonged viral shedding. We performed whole-genome sequencing (WGS) of SARS-CoV-2 to evaluate the possibility of post-isolation transmission from this prolonged viral shedding.
Results
A patient (case 1) with acute myeloid leukemia was released from isolation 54 days after the diagnosis of COVID-19 based on rising Ct value of up to 29.3, and moved to a six-patient room. On days 10 and 11 post-isolation, his doctor (case 2) and 2 patients who were his roommates (case 3, 4) had positive SARS-CoV-2 PCR results. Additionally, 16 days post-isolation, another patient (case 5) in a remote room had positive SARS-CoV-2 PCR result. All the three patients were hospitalized for ≥ 14 days when they were diagnosed with SARS-CoV-2 infection. Except for case 3, the remaining 4 cases were available for WGS, which revealed that case 1 exhibited a 7 nucleotides difference in comparison to cases 4 and 5 and case 2 displayed a 20 nucleotides difference compared with case 1, while sequences of cases 4 and 5 were identical.
Conclusions
Despite the possibility of transmission from the patient with prolonged viral shedding, no evidence of the transmission of SARS-CoV-2 from the patient with prolonged positive RT-PCR using WGS was found.
Go to :

Graphical Abstract
Go to :

Keywords: SARS-CoV-2, COVID-19, Whole-Genome Sequencing, Hematologic Malignancy, Post-Isolation Transmission
INTRODUCTION
As moderately or severely immunocompromised patients may shed replication-competent virus more than 20 days post-infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), Centers for Disease Control and Prevention (CDC) recommends an isolation period of at least 20 days with the use of test-based strategy and consultation with an infectious disease specialist to determine the duration of isolation.
1 However, patients with hematologic malignancies often have prolonged positive real-time polymerase chain reaction (RT-PCR) results after an initially positive viral test.
23 The culture-based isolation of virus may determine whether a positive RT-PCR result contains viable virus; however, this is labor intensive and time consuming.
4 Moreover, using test-based strategy, isolation period is lengthened and adequate treatment for underlying hematologic malignancy might be delayed when patients are isolated in designated coronavirus disease 2019 (COVID-19) treatment units. In addition, the data regarding transmission of SARS-CoV-2 from patients with prolonged viral shedding is limited.
During the era of the dominance of the omicron variant (May 2022), nosocomial clusters of COVID-19 were detected in the hematologic ward, post-isolation of the patient (case 1) with prolonged positive RT-PCR results. Four additional cases included 2 patients from the same room as the index case, the physician attending the index case, and 1 case from another room. Herein, we report the results of an epidemiologic investigation using whole-genome sequencing (WGS), performed to determine the transmission from case 1 to other cases.
Go to :

METHODS
Setting
A COVID-19 cluster occurred in the hematologic ward of Asan Medical Center, a 2,700-bed tertiary care center in Seoul, Korea, during May 2022. The ward has 19 rooms with 4 six-patient rooms, a 1 five-patient room, 7 two-patient rooms, and 7 single-patient rooms. Adult patients with hematologic malignancies and hematopoietic stem cell transplant recipients stayed in this ward. Before admission, all patients were screened for SARS-CoV-2 using RT-PCR, and were tested again during their stay (at day 3 and 7) regardless of symptoms. During hospitalization, SARS-CoV-2 RT-PCR test was performed whenever patients developed symptoms of COVID-19. Only one resident caregiver per patient was permitted, and entry into this ward was strictly limited to registered caregivers and healthcare workers.
Contact tracing was performed when patients, caregivers, or healthcare workers were confirmed with COVID-19. For inpatients and their caregiver diagnosed with COVID-19, their roommates were classified as being in close contact and isolated in a single room. For healthcare workers, close contact was defined as being within 2 meters of cases not wearing a mask for more than 3 minutes without wearing both mask and face shield (or goggles). They performed testing on the day of the index patient’s diagnosis and on the 2nd and 5th days. Universal masking was mandated in the hospital, while the healthcare workers wore KF94 (FFP2 equivalent) mask and goggles or face shield.
WGS
To analyze viral genomic sequences from the index sample, viral RNA was extracted using a QIAamp viral RNA Mini Kit (Qiagen, Hilden, Germany). To isolate pure SARS-CoV-2 RNA, human ribosomal RNA was depleted using the NEBNext rRNA Depletion Kit (New England Biolabs, Ipswich, MA, USA). Libraries were prepared using the TruSeq RNA sample prep kit v2 (Illumina, San Diego, CA, USA) protocol. The enriched libraries were quantified using the Kapa Library Quantification Kit (Roche, Basel, Switzerland), and the sequencing was performed using the Miseq platform (Illumina) with Miseq reagent kit v2 (300 cycles) (Illumina). Sequencing analysis was performed using CLC Genomics Workbench 10 (Qiagen). Base-called reads in FASTQ were trimmed and mapped to a reference sequence (NCBI Reference: NC_045512). WGS data > 2 single-nucleotide polymorphisms apart were deemed to not be cluster related.
5
Sequence analysis
PANGO lineages of genomic sequences acquired from the 4 cases were determined using Phylogenetic Assignment of Named Global Outbreak LINeages version 4.1.1 (
https://pangolin.cog-uk.io; last accessed August 4). To analyze the mutation frequencies, full-length and high coverage sequences of SARS-CoV-2 omicron variants (n = 536) isolated in May 2022 in South Korea were retrieved from the Global Initiative on Sharing All Influenza Data (GISAID) database (accessed August 2). Nucleotide sequences were aligned to the reference genome (NCBI Reference: NC_045512) using MAFFT (version 7.471). After alignment, we analyzed the mutation proportion at specific nucleotide sites.
Ethics statement
The Institutional Review Board of Asan Medical Center evaluated and approved the medical, scientific, and ethical aspects of the study protocol and the requirement for informed consent was waived (IRB No. 2021-0024).
Go to :

RESULTS
Description of cases
A 59-year-old man (case 1) with relapsing acute myeloid leukemia after hematopoietic stem cell transplant was diagnosed with COVID-19 on March 10, 2022 in the community. He was admitted to the designated COVID-19 isolation ward on April 5 with the diagnosis of COVID-19 pneumonia because of fever, cough, and sputum. Upon admission, cycle threshold (Ct) value of
E gene from a nasopharyngeal swab was 17.36. With O2 requirement (3L/min via nasal prong), he was treated with remdesivir for 5 days and dexamethasone for 6 days. Fifteen days post-admission (April 20), he was transferred to the hematologic ward as his condition improved, and he was isolated in a single room in the ward. On April 26, chemotherapy with venetoclax and decitabine was started. After consultation with an infectious disease specialist (S-H Kim), he was released from isolation 54 days after the diagnosis of COVID-19 (May 3) based on rising Ct value of up to 29.3, and moved to a six-patient room (
Fig. 1). From 10 to 11 days post-isolation (May 13 and 14), his doctor (case 2) and his 2 roommates (case 3, 4) had positive SARS-CoV-2 PCR results. Case 2 was casual contact to case 1 and cases 3, 4 were close contact as they were roommates with case 1. PCR testing was performed for cases 2 and 3 owing to symptoms and for case 4 owing to contact with case 3. In addition, 16 days post-isolation, another patient (case 5) in a remote room had positive SARS-CoV-2 PCR result, which was performed owing to fever. All three patients were hospitalized for ≥ 14 days when they were diagnosed with COVID-19. On May 15, Ct value of SARS-CoV-2 PCR result of case 1 was 25.79. Demographic characteristics of cases are given in
Table 1.
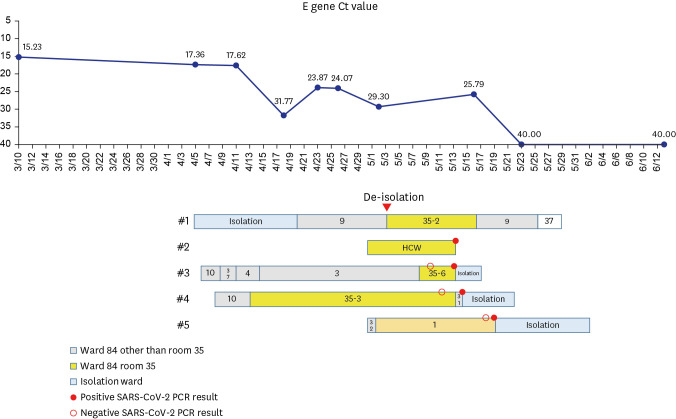 | Fig. 1
Ct value (E gene, case 1), diagnosis day and spatiotemporal relationship between cases.
Ct = cycle threshold, HCW = healthcare worker, SARS-CoV-2 = severe acute respiratory syndrome coronavirus 2, PCR = polymerase chain reaction.

|
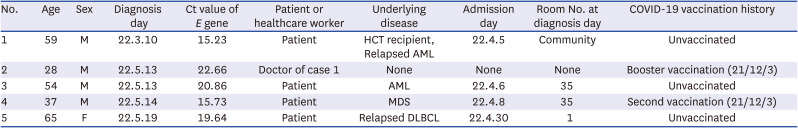
Table 1
Demographic characteristics of the cases
|
No. |
Age |
Sex |
Diagnosis day |
Ct value of E gene |
Patient or healthcare worker |
Underlying disease |
Admission day |
Room No. at diagnosis day |
COVID-19 vaccination history |
|
1 |
59 |
M |
22.3.10 |
15.23 |
Patient |
HCT recipient, Relapsed AML |
22.4.5 |
Community |
Unvaccinated |
|
2 |
28 |
M |
22.5.13 |
22.66 |
Doctor of case 1 |
None |
None |
None |
Booster vaccination (21/12/3) |
|
3 |
54 |
M |
22.5.13 |
20.86 |
Patient |
AML |
22.4.6 |
35 |
Unvaccinated |
|
4 |
37 |
M |
22.5.14 |
15.73 |
Patient |
MDS |
22.4.8 |
35 |
Second vaccination (21/12/3) |
|
5 |
65 |
F |
22.5.19 |
19.64 |
Patient |
Relapsed DLBCL |
22.4.30 |
1 |
Unvaccinated |

WGS analysis
Except for case 3, other 4 cases underwent WGS. Nasopharyngeal swabs for WGS from case 1 and 4 were obtained on May 18, and the specimen from case 2 and that from case 5 were obtained on May 19 and on May 20, respectively. Sequences of cases 4 and 5 were identical, while case 1 exhibited a 7 nucleotides difference (5 amino acid differences) in comparison to cases 4 and 5 (
Fig. 2). The differences that were observed included a 2 nucleotides difference in ORF1ab (3965 and 3971), 3 nucleotides deletion and 1 nucleotide difference in Spike (430–432 and 433), and 1 nucleotide difference in N (53) genes. These sites are known to be highly conserved at South Korean omicron isolates (isolated in May 2022, n = 536), and no identical sequences found in cases 1, 4, and 5 have been further identified. On the other hand, case 2 (solely assigned as BA.2.3) displayed a 20 nucleotides difference (15 amino acid differences) compared with case 1.
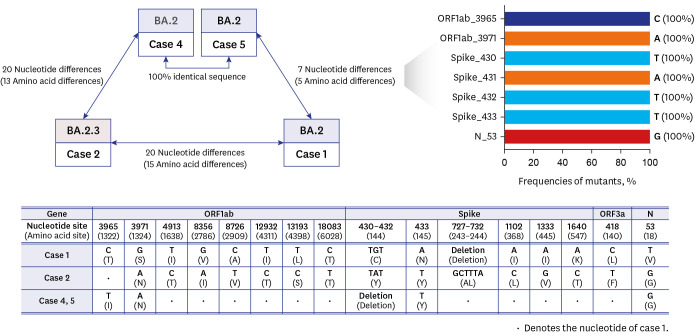 | Fig. 2Comparison of whole-genome sequences of the cases.
|
Go to :

DISCUSSION
Although viable virus can be isolated up to 61 days post-onset of symptoms in immunocompromised patients,
2 the data regarding the true transmission from patients with prolonged viral shedding and contact > 10 days post-onset of their symptoms is limited. During the omicron surge, it was difficult to isolate the patients with hematologic malignancies using test-based strategy (isolation until two consecutive negative results), especially in the ward with multi-patient rooms and limited number of single-patient isolation room. Therefore, in case 1, the patient was deisolated 8 weeks post-diagnosis considering the rise in Ct value up to 29. As two roommates were confirmed with COVID-19 ten days after sharing the room, there were concerns regarding transmission from case 1 to other two patients. Our experience proved that it is difficult to determine the source of infection solely based on the epidemiologic association during the omicron surge, as there could be multiple sources of infection. Additionally, WGS analysis is a valuable tool to identify the source of infection. Through WGS analysis, no evidence of transmission from the patient with prolonged positive RT-PCR results who was released from isolation about 8 weeks after diagnosis was found. Therefore, conventional epidemiologic investigations suggesting the transmission from those with prolonged viral shedding or post-isolation transmission might be overestimated. Hence, WGS data would provide the exact evaluation of transmission dynamics.
6789
The specimen for WGS from case 1 was obtained at day 69 after his diagnosis of COVID-19 when the cluster was occurred. Although it is possible that intrahost mutation could be occurred especially in immunocompromised patients, we believe that it is less likely that 7 nucleotide mutation occurred during transmission from case 1 to cases 3 and 4. First, the specimens were obtained simultaneously. Second, considering that previous studies showed that 3-point mutation occurred 76 days after diagnosis, and 1 point mutation occurred 73 days after diagnosis,
1011 we believe that 7 mutation occurred during 69 days indicates too rapid evolution.
This study has certain limitations. First, we could not perform a thorough epidemiologic investigation due to limited resource after the omicron surge, and we did not find other sources of infection for cases 2–5. Second, culture for virus isolation for case 1 was not performed.
In conclusion, our study suggests that it is difficult to determine the source of infection solely based on epidemiologic link during endemic period, and WGS is valuable tool to determine it.
Go to :







 PDF
PDF Citation
Citation Print
Print




 XML Download
XML Download