Abstract
Skeletal muscle is now regarded as an endocrine organ based on its secretion of myokines and exerkines, which, in response to metabolic stimuli, regulate the crosstalk between the skeletal muscle and other metabolic organs in terms of systemic energy homeostasis. This conceptual basis of skeletal muscle as a metabolically active organ has provided insights into the potential role of physical inactivity and conditions altering muscle quality and quantity in the development of multiple metabolic disorders, including insulin resistance, obesity, and diabetes. Therefore, it is important to understand human muscle physiology more deeply in relation to the pathophysiology of metabolic diseases. Since monolayer cell lines or animal models used in conventional research differ from the pathophysiological features of the human body, there is increasing need for more physiologically relevant in vitro models of human skeletal muscle. Here, we introduce recent studies on in vitro models of human skeletal muscle generated from adult myogenic progenitors or pluripotent stem cells and summarize recent progress in the development of three-dimensional (3D) bioartificial muscle, which mimics the physiological complexity of native skeletal muscle tissue in terms of maturation and functionality. We then discuss the future of skeletal muscle 3D-organoid culture technology in the field of metabolic research for studying pathological mechanisms and developing personalized therapeutic strategies.
Metabolic pathways regulate and maintain balance in the processing and distribution of nutrients. This involves complex networks and communication between multiple metabolic organ systems working together for the efficient conversion of chemical energy, and any imbalance leading to dysregulated metabolic processes leads to several metabolic disorders characterized by increased insulin resistance, dyslipidemia, obesity, and diabetes. As a crucial metabolic regulator, skeletal muscle is an important target for understanding and developing treatment interventions for metabolic diseases. Skeletal muscle is the primary site of insulin-mediated glucose disposal and the storage of glycogen and amino acids; furthermore, it modulates the catabolism of circulating and stored lipids [1]. Beyond energy consumption, skeletal muscle activity induces changes in metabolic processes through the browning of white adipose tissue and by increasing insulin sensitivity through exercise and muscle contraction [2]. Structurally, skeletal muscles are composed of bundles of muscle fibers surrounded by connective tissue. They are nutritionally supported by blood vessels and produce voluntary movement upon stimulation by motor neurons [3].
Although rodent models are a common tool for metabolic disease research, research, clinical translation to human patients is limited due to the substantial physiological differences between rodents and humans [4,5]. Furthermore, animal models that better model the pathophysiology of humans, such as nonhuman primates, are limited in their potential for genetic engineering, costs, and handleability [6]. Thus, human stem cell-based models have received widespread attention in metabolic disease research. While human-derived skeletal muscle models can overcome the practical and translational limitations of animal models, the conventional monolayer culture method of muscle stem cells does not generate the vascular networks and interactions with nerve fibers observed in vivo. However, with the advent of in vitro human skeletal muscle (hSkM) that adds multicellular complexity, it is now possible to address the limitations of traditional culture systems. In this review, we highlight advanced three-dimensional (3D) models of bioartificial hSkM using adult myogenic progenitors or pluripotent stem cells (PSCs) and discuss potential applications for the progress of metabolic disease research. We hope that this information will help many researchers who are considering using novel systems to study human metabolic diseases.
Biomedical research has traditionally focused on a handful of model organisms based on the consensus that essential biological mechanisms are conserved throughout species [7]. Among mammalian model systems, the mouse is the preferred research model due to the ease of genetic engineering [8,9]. Although many animal models have provided important insights into skeletal muscle development and pathology, the etiology and pathophysiology of metabolic diseases, genetic variation within the human population, and certain drug responses are not recapitulated in animal models, necessitating the development of human-specific model systems [5,10].
Bioartificial hSkM is a source of implantable tissue for regenerative medicine and has been an attractive research tool for studying muscle disorders and regeneration. There are two primary approaches to hSkM production (Fig. 1): (1) utilizing adult muscle stem cells derived from patient tissue and (2) inducing differentiation of induced pluripotent stem cells (iPSCs) into muscle stem cells. These methods enable artificial skeletal muscle models to mimic human muscle development and regeneration, providing valuable information about the underlying mechanisms, and have potential for applications in regenerative and metabolic research.
Primary muscle stem cells isolated directly from patient tissue have been predominantly used for modeling skeletal muscle function and disease due to existing knowledge and established methods of muscle stem cell isolation and culture conditions [11,12]. However, hSkM derived from adult muscle stem cells is limited in its differentiation capacity, cell expansion potential, and the availability of human samples [13-15]. Furthermore, as it lacks the surrounding microenvironment such as blood vessels, neuromuscular junctions, and fibro-adipogenic progenitors, hSkM derived from adult stem cells cannot fully reflect in vivo conditions [3,13,16-18]. Therefore, PSCs, including iPSCs and embryonic stem cells, may be more suitable for advanced in vitro models of metabolic diseases.
The introduction of four transcription factors (Oct4, Sox2, c-Myc, and Klf4) via retroviral constructs can reprogram somatic cells such as fibroblasts, peripheral blood mononuclear cells, and core blood mononuclear cells into iPSCs, which have the capacity for self-renewal and differentiation into muscle progenitor cells. The development of iPSC-derived hSkM technology has led to fundamental changes in the use of stem cells in disease modeling, circumventing ethical concerns about the use of embryonic stem cells and reducing the need for invasive muscle biopsies in hSkM research. A comprehensive understanding of embryonic myogenesis by myogenic regulatory factors (MRFs) and signaling molecules has been conducive to building muscle tissue from PSCs. During the embryonic stage, skeletal muscles are derived from the dermomyotome, which originates from the paraxial mesoderm [19]. Muscle progenitor cells delaminate from the edges of the dermomyotome under the regulation of MRFs such as paired box 7 (PAX7), myogenic factor 5 (MYF5), myoblast determination protein (MYOD), myogenin (MYOG), and myogenic regulatory factor 4 (MRF4) and commence myogenesis [20]. Simultaneously, as key signaling pathways, Wnt, Notch, Sonic hedgehog, and bone morphogenetic proteins are spatiotemporally activated to contribute to this myogenic program [21]. The committed myoblast expressing PAX3/7 and MYOD extensively proliferates for myonuclear accretion into the myotube. Eventually, these myotubes are further fused into mature myofibers. During puberty, a portion of the muscle stem cells exit the cell cycle and become adult muscle stem cells, which are also known as satellite cells [21,22]. Adult muscle stem cells are located between the basal lamina and sarcolemma of myofibers and are among the most well-defined adult stem cells. Upon injury, these stem cells exit quiescence and divide into a pool of myogenic progenitors to rebuild damaged myofibers [23].
In the past several years, researchers have established various culture conditions based on a detailed understanding of myogenic development and have found appropriate combinations of essential factors to differentiate myogenic cells from PSCs. In-depth reviews have been published by Jalal et al. [13] and Iberite et al. [24]. Chiefly, there are two strategies for inducing differentiation from human PSCs. The first is to induce differentiation by exogenous expression of transcriptional regulators such as PAX7, MYOD, and MYF5. The second is to use a combination of signaling molecules, growth factors, and inhibitors without the overexpression of exogenous genes.
Under optimized iPSC culture conditions, Maffioletti et al. [25] generated a patient-specific iPSC-derived muscle model that contained the major cellular components of skeletal muscle, including vascular endothelial cells, pericytes, and motor neurons. These results lay the foundation for a hSkM organoid-like platform for disease modeling, regenerative medicine, and therapeutic development.
Furthermore, Rao et al. [26] developed a method of 3D contractile skeletal muscle tissues derived from human PSCs transduced with lentiviruses encoding doxycycline-inducible expression of Pax7. When cultured in a 3D hydrogel environment, the functional skeletal muscle tissues were able to generate contractions and Ca2+ transients in response to electrical and neurotransmitter stimulation [26].
We discuss the 3D approach in more detail below. Although iPSCs can generate hSkM cells, there remain questions regarding how well the in vitro characterization of these cells recapitulates the in vivo biology and processes seen in metabolic diseases. If disease-associated phenotypes can be established from iPSC-derived hSkM cells, this would open the door to understanding the complex nature of metabolic disease.
In this regard, a recent study showed promising results in elucidating the physiological consequences of type 2 diabetes with an in vitro disease-in-a-dish model using iPSCs from type 2 diabetes patients. After differentiating patient-derived iPSCs into myoblasts, the resulting cells exhibited multiple defects mirroring human disease, including insulin resistance, reduced insulin-stimulated glucose uptake, and reduced mitochondrial oxidation [27].
Although two-dimensional (2D) myofiber culture systems have been widely used in muscle disease research, they do not recapitulate the complexity of the functional units and niches in the muscle tissue. Therefore, a strategy has been established to mimic the complex 3D structure of skeletal muscle using biomaterial scaffolds and specific biofabrication techniques to engineer skeletal muscle in vitro.
The advancement of hSkM tissue models require consideration of structural, operational, and environmental factors to recapitulate primary tissues. Broadly, 3D culture systems have three key advantages over 2D cultures: the 3D architecture, improved maturation and functionality, and the multi-lineage complexity.
The generation of 3D skeletal muscles can be categorized into organoid-like self-assembling and scaffold-based methods. Organoids serve as a powerful model that reproduces the cellular heterogeneity and function of primary tissues in addition to the ability to differentiate and self-organize. While muscle organoids can model the complexity of muscle tissue and cellular heterogeneity, organoid growth conditions cannot recreate myofiber structures due to the lack of the force and tension that naturally occur with muscle contraction and anchorage to the bone. Scaffold-based 3D culture models overcome this limitation by providing structural support and mechanical cues to mimic the physiological conditions. One method of carrying this out is by embedding differentiated human myoblasts in hydrogels and attaching them between two points, generating tension in the hydrogel and aligning myofibers along the central axis. This method is advantageous for sarcomere maturation and has been useful for revealing disease phenotypes that affect skeletal muscle structure [25,28,29].
While 3D culture models have the advantage of improved structural organization, they lack the multicellular complexity of muscle tissue in vivo due to their single-lineage origin. This limitation has been addressed by including cells of multiple lineages, such as endothelial cells and pericytes along with myogenic cells. This approach has shown signs of improved recovery upon injury after implantation in mice and the development of vascular-like networks [25]. In addition, myogenic progenitors can be differentiated in hydrogels before being embedded in a hydrogel containing endothelial cells and myofibroblasts. This method has been useful for demonstrating the heterotypic interactions between muscle, endothelial, and fibroblast cells in a model of Duchenne muscular dystrophy that could not be observed in a 2D model, including an increase in the expression of fibrosis-associated proteins with the incorporation of patient-derived fibroblasts [30].
Integrating the 3D structure and lineage complexity of muscle tissue, bioprinting techniques enabled the controlled spatial patterning of cells. This technique utilizes bioink loaded with biomaterials and myogenic progenitors to align the cells into multilayered strips for precise alignment and formation of a 3D structure. The addition of bioink containing neural progenitors and endothelial cells further improves muscle function, neuromuscular junction formation, and vascularization upon implantation [31].
While recapitulating the 3D structure and complexity are important for modeling the skeletal muscle tissue, enhancing the function and simulating innervation contribute to generating a more comprehensive model. Muscle function is controlled by motor neurons and the release of acetylcholine at the neuromuscular junction that triggers an action potential in the muscle. Muscle innervation and muscle contraction can be simulated in vitro by chemical or electrical stimulation, as well as optogenetic control [14,17,25], resulting in hypertrophy, myosin isoform switching, and increased force [32]. Employing these methods, Osaki et al. [17] demonstrated degradation of motor neurons and a decline in muscle function in a microfluidic device of 3D skeletal muscle innervated by iPSC-derived motor neuron spheroids from an amyotrophic lateral sclerosis (ALS) patient.
One of the challenges of generating a hSkM model of metabolic disease relates to the complexity of metabolic diseases. While skeletal muscle makes a direct, active contribution to metabolic diseases in its role as an endocrine organ that regulates energy metabolism and as a major site of insulin-mediated glucose [1], it is also adversely affected by metabolic disease [33]. While it is well-established that type 2 diabetes is linked to motor dysfunction, microvascular dysfunction, and neuropathy, there has been less focus on the inter-related effects between muscle dysfunction and peripheral tissues, potentially due to the unavailability of a sufficient human model of metabolic diseases. This underscores the value of an advanced model of hSkM that can integrate innervation and vasculature into the system.
Diabetes causes a variety of medical complications associated with metabolic diseases. Impairments of micro-vessels caused by diabetes result in nephropathy, retinopathy, and diabetic neuropathy. Type 2 diabetes mellitus is also associated with degeneration of the skeletal muscle microenvironment [34,35]. These wide-ranging complications have become a major bottleneck in the understanding of pathological mechanisms and the drug discovery process due to the lack of accurate models that mimic the pathology. The emergence of human in vitro 3D models using stem cells from different organs, including skeletal muscle, has received much attention for the potential to address this limitation [14,25,26].
Previously, PSC-derived hSkM has been primarily used in research on muscle diseases, such as ALS and Duchenne muscular dystrophy, due to the ability of this method to precisely recapitulate patients’ genetic background [17,25,28,36]. With recent advances in hSkM production and gene engineering using CRISPR-Cas systems, the application of PSC-derived hSkM is expected to expand to metabolic disease research in the discovery of new disease mechanisms and provide better reliability than existing 2D models for drug development. Although not yet attempted in 3D hSkM models, muscle fiber cultures derived from patients with Donohue syndrome have successfully reproduced the deficiencies in insulin signaling, glucose uptake, glycogen accumulation, and insulin-regulated gene expression that are characteristic of the syndrome [37]. In the future, understanding the progression and consequences of insulin resistance, as well as the vascular damage and consequent diabetic neuropathy, will be important for drug development.
Thus far, appropriate model platforms have been limited by inadequate translatability and incomplete reproduction of the native microenvironment of the skeletal muscle tissue. Furthermore, the property of metabolic diseases as a cluster of conditions that affect and are influenced by communication between multiple distal tissues beyond the local microenvironment further complicates the challenge of creating a human-based model of metabolic diseases. While current models of hSkM are limited in representing the complex disease networks of metabolic diseases, strides have been made in the development of advanced culture systems and bioengineering techniques that will propel metabolic research and precision medicine (Fig. 2). Although skeletal muscle is a central site for energy consumption, it also plays an active role in modulating energy metabolism as an endocrine organ that communicates with multiple organ systems by secreted factors called myokines and exerkines [38]. Moving forward, we see a need for better ways to model these inter-organ communication networks observed in metabolic diseases to better understand the pathophysiology behind metabolic disorders and develop treatment interventions tailored to the patient.
ACKNOWLEDGMENTS
This work has supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (NRF-2022R1C1C1006015, NRF-2021RIF1A1061077).
REFERENCES
1. Smith AG, Muscat GE. Skeletal muscle and nuclear hormone receptors: implications for cardiovascular and metabolic disease. Int J Biochem Cell Biol. 2005; 37:2047–63.

2. Otero-Diaz B, Rodriguez-Flores M, Sanchez-Munoz V, Monraz-Preciado F, Ordonez-Ortega S, Becerril-Elias V, et al. Exercise induces white adipose tissue browning across the weight spectrum in humans. Front Physiol. 2018; 9:1781.
3. Frontera WR, Ochala J. Skeletal muscle: a brief review of structure and function. Calcif Tissue Int. 2015; 96:183–95.

4. Doncheva NT, Palasca O, Yarani R, Litman T, Anthon C, Groenen MA, et al. Human pathways in animal models: possibilities and limitations. Nucleic Acids Res. 2021; 49:1859–71.

5. Sanoh S, Horiguchi A, Sugihara K, Kotake Y, Tayama Y, Uramaru N, et al. Predictability of metabolism of ibuprofen and naproxen using chimeric mice with human hepatocytes. Drug Metab Dispos. 2012; 40:2267–72.

6. Varga O, Harangi M, Olsson IA, Hansen AK. Contribution of animal models to the understanding of the metabolic syndrome: a systematic overview. Obes Rev. 2010; 11:792–807.

7. Kafkafi N, Agassi J, Chesler EJ, Crabbe JC, Crusio WE, Eilam D, et al. Reproducibility and replicability of rodent phenotyping in preclinical studies. Neurosci Biobehav Rev. 2018; 87:218–32.

8. Bruning JC, Michael MD, Winnay JN, Hayashi T, Horsch D, Accili D, et al. A muscle-specific insulin receptor knockout exhibits features of the metabolic syndrome of NIDDM without altering glucose tolerance. Mol Cell. 1998; 2:559–69.

9. Wang B, Chandrasekera PC, Pippin JJ. Leptin- and leptin receptor-deficient rodent models: relevance for human type 2 diabetes. Curr Diabetes Rev. 2014; 10:131–45.

10. Cox TC. Utility and limitations of animal models for the functional validation of human sequence variants. Mol Genet Genomic Med. 2015; 3:375–82.

11. Xu X, Wilschut KJ, Kouklis G, Tian H, Hesse R, Garland C, et al. Human satellite cell transplantation and regeneration from diverse skeletal muscles. Stem Cell Reports. 2015; 5:419–34.

12. Garcia SM, Tamaki S, Lee S, Wong A, Jose A, Dreux J, et al. High-yield purification, preservation, and serial transplantation of human satellite cells. Stem Cell Reports. 2018; 10:1160–74.

13. Jalal S, Dastidar S, Tedesco FS. Advanced models of human skeletal muscle differentiation, development and disease: three-dimensional cultures, organoids and beyond. Curr Opin Cell Biol. 2021; 73:92–104.

14. Afshar Bakooshli M, Lippmann ES, Mulcahy B, Iyer N, Nguyen CT, Tung K, et al. A 3D culture model of innervated human skeletal muscle enables studies of the adult neuromuscular junction. Elife. 2019; 8:e44530.

15. Kim J, Koo BK, Knoblich JA. Human organoids: model systems for human biology and medicine. Nat Rev Mol Cell Biol. 2020; 21:571–84.

16. Moyle LA, Jacques E, Gilbert PM. Engineering the next generation of human skeletal muscle models: from cellular complexity to disease modeling. Curr Opin Biomed Eng. 2020; 16:9–18.

17. Osaki T, Uzel SG, Kamm RD. Microphysiological 3D model of amyotrophic lateral sclerosis (ALS) from human iPS-derived muscle cells and optogenetic motor neurons. Sci Adv. 2018; 4:eaat5847.

18. Biferali B, Proietti D, Mozzetta C, Madaro L. Fibro-adipogenic progenitors cross-talk in skeletal muscle: the social network. Front Physiol. 2019; 10:1074.

19. Hernandez-Hernandez JM, Garcia-Gonzalez EG, Brun CE, Rudnicki MA. The myogenic regulatory factors, determinants of muscle development, cell identity and regeneration. Semin Cell Dev Biol. 2017; 72:10–8.

20. Bentzinger CF, Wang YX, Rudnicki MA. Building muscle: molecular regulation of myogenesis. Cold Spring Harb Perspect Biol. 2012; 4:a008342.

21. Tajbakhsh S. Skeletal muscle stem cells in developmental versus regenerative myogenesis. J Intern Med. 2009; 266:372–89.

22. Kim JH, Han GC, Seo JY, Park I, Park W, Jeong HW, et al. Sex hormones establish a reserve pool of adult muscle stem cells. Nat Cell Biol. 2016; 18:930–40.

23. Tedesco FS, Dellavalle A, Diaz-Manera J, Messina G, Cossu G. Repairing skeletal muscle: regenerative potential of skeletal muscle stem cells. J Clin Invest. 2010; 120:11–9.

24. Iberite F, Gruppioni E, Ricotti L. Skeletal muscle differentiation of human iPSCs meets bioengineering strategies: perspectives and challenges. NPJ Regen Med. 2022; 7:23.

25. Maffioletti SM, Sarcar S, Henderson AB, Mannhardt I, Pinton L, Moyle LA, et al. Three-dimensional human iPSC-derived artificial skeletal muscles model muscular dystrophies and enable multilineage tissue engineering. Cell Rep. 2018; 23:899–908.

26. Rao L, Qian Y, Khodabukus A, Ribar T, Bursac N. Engineering human pluripotent stem cells into a functional skeletal muscle tissue. Nat Commun. 2018; 9:126.

27. Batista TM, Jayavelu AK, Wewer Albrechtsen NJ, Iovino S, Lebastchi J, Pan H, et al. A cell-autonomous signature of dysregulated protein phosphorylation underlies muscle insulin resistance in type 2 diabetes. Cell Metab. 2020; 32:844–59.

28. Ebrahimi M, Lad H, Fusto A, Tiper Y, Datye A, Nguyen CT, et al. De novo revertant fiber formation and therapy testing in a 3D culture model of Duchenne muscular dystrophy skeletal muscle. Acta Biomater. 2021; 132:227–44.

29. Rajabian N, Shahini A, Asmani M, Vydiam K, Choudhury D, Nguyen T, et al. Bioengineered skeletal muscle as a model of muscle aging and regeneration. Tissue Eng Part A. 2021; 27:74–86.

30. Bersini S, Gilardi M, Ugolini GS, Sansoni V, Talo G, Perego S, et al. Engineering an environment for the study of fibrosis: a 3D human muscle model with endothelium specificity and endomysium. Cell Rep. 2018; 25:3858–68.

31. Choi YJ, Jun YJ, Kim DY, Yi HG, Chae SH, Kang J, et al. A 3D cell printed muscle construct with tissue-derived bioink for the treatment of volumetric muscle loss. Biomaterials. 2019; 206:160–9.

32. Wang J, Khodabukus A, Rao L, Vandusen K, Abutaleb N, Bursac N. Engineered skeletal muscles for disease modeling and drug discovery. Biomaterials. 2019; 221:119416.

33. Nawrocki AR, Scherer PE. The delicate balance between fat and muscle: adipokines in metabolic disease and musculoskeletal inflammation. Curr Opin Pharmacol. 2004; 4:281–9.

35. Teng S, Huang P. The effect of type 2 diabetes mellitus and obesity on muscle progenitor cell function. Stem Cell Res Ther. 2019; 10:103.

36. Faustino Martins JM, Fischer C, Urzi A, Vidal R, Kunz S, Ruffault PL, et al. Self-organizing 3D human trunk neuromuscular organoids. Cell Stem Cell. 2020; 27:498.

Fig. 1.
Recreation of human skeletal muscle in vitro. Human skeletal muscle can be established in vitro using induced pluripotent stem cells (iPSCs) and adult muscle stem cells (MuSCs). iPSCs are differentiated by the exogenous expression of transcriptional regulators or by using a combination of growth factors and signaling molecules. The resulting myoblasts may be encapsulated in support substrates such as hydrogels and formed into myotubes, then arranged into three-dimensional (3D) myofibers mimicking the skeletal muscle tissue. iPSC-derived neural and endothelial cells may be included along with supporting cells to form more complex models that better recapitulate the microenvironment of native skeletal muscles. 3D bioprinting and scaffold-based culture methods may be used for improved replication of native muscle tissue structure, and muscle innervation may be simulated to test the function and performance of the constructed skeletal muscle. BMP, bone morphogenic protein; Shh, sonic hedgehog protein; Pax7, paired box 7; MyoD, myoblast determination protein; Myf5, myogenic factor 5; PSC, pluripotent stem cell; Cxcl4, CXC chemokine ligand 4; 2D, three-dimensional.
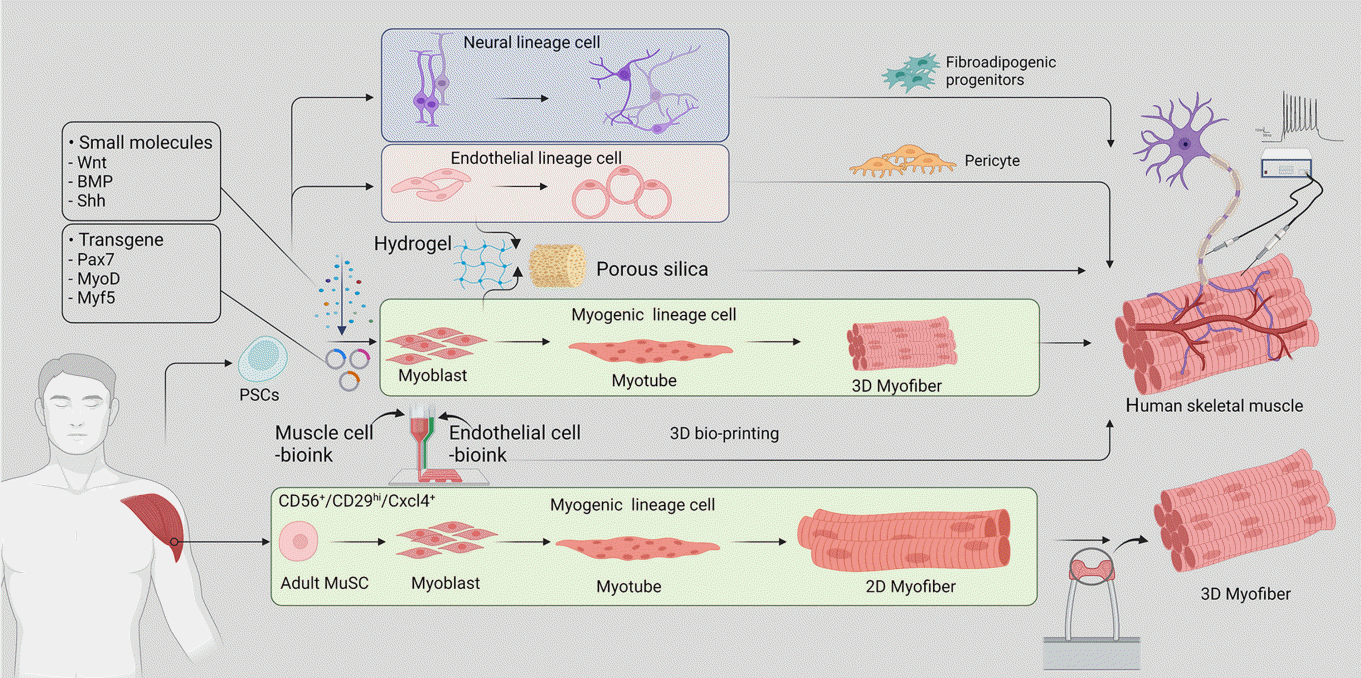
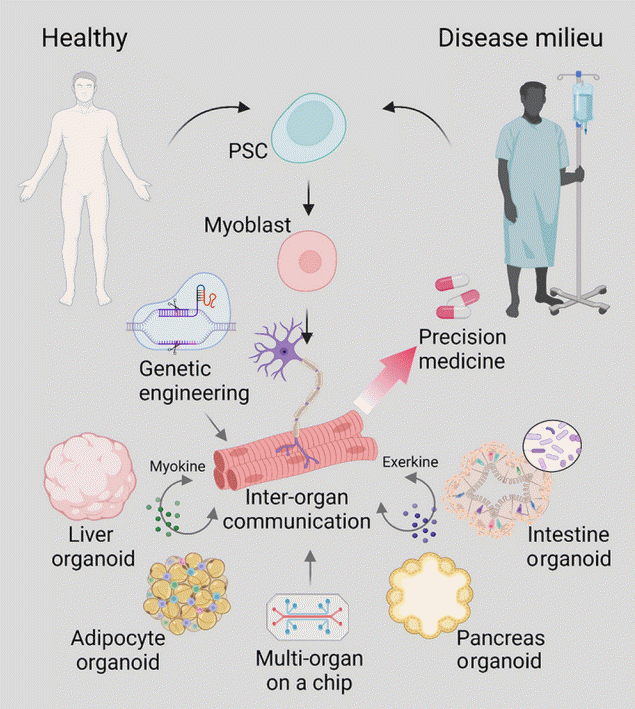
Fig. 2.
Potential applications of three-dimensional human skeletal muscle. The skeletal muscle is a major regulator of energy metabolism that communicates with multiple organ systems and is one of the major organs affected by metabolic syndrome. Devices such as multi-organ on a chip can be used to replicate the inter-organ communication between the skeletal muscle and other organs such as the liver, serving as a platform to comprehensively model these communication networks. Furthermore, recent advances in genetic engineering technology and cell reprogramming using patient-derived pluripotent stem cells (PSCs) have made it possible to generate disease models of metabolic syndrome, facilitating the path toward precision medicine.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download