Abstract
Background
Methods
Results
Notes
ACKNOWLEDGMENTS
REFERENCES
Fig. 1.
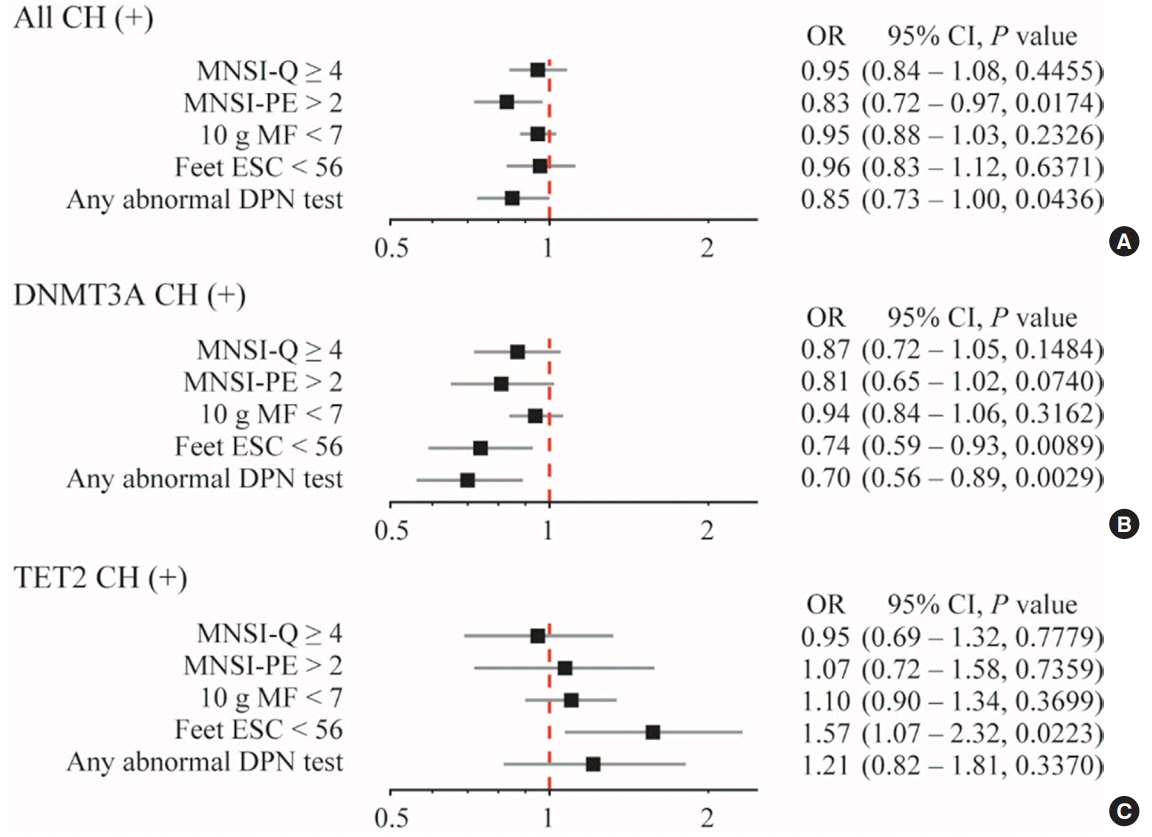
Table 1.
| Characteristic | DPN (–) (n=181) | DPN (+) (n=113) | P valuea |
|---|---|---|---|
| Age, yr | 58.7±9.4 | 59.7±10.1 | 0.265 |
| Male sex | 117 (64.6) | 67 (59.3) | 0.387 |
| Height, cm | 164.2±8.8 | 163.5±9.2 | 0.530 |
| Body weight, kg | 67.4±11.2 | 69.6±12.7 | 0.120 |
| BMI, kg/m2 | 24.9±3.2 | 26.0±3.7 | 0.009 |
| Waist circumference, cm | 87.9±8.7 | 89.8±9.1 | 0.072 |
| SBP, mm Hg | 130±14 | 131±15 | 0.677 |
| DBP, mm Hg | 75±10 | 75±10 | 0.535 |
| Diabetes duration, yr | 10.8±8.3 | 12.0±8.3 | 0.162 |
| FPG, mmol/L | 7.5±1.7 | 8.0±2.8 | 0.215 |
| HbA1c, mmol/mmol | 54±12.0 | 58±15.3 | 0.015 |
| HOMA-IR | 2.6±1.4 | 3.0±1.8 | 0.236 |
| Cholesterol, mmol/L | 4.1±1.0 | 3.9±1.0 | 0.060 |
| Triglyceride, mmol/L | 1.44±0.94 | 1.65±1.58 | 0.652 |
| HDL-C, mmol/L | 1.30±0.34 | 1.20±0.31 | 0.013 |
| LDL-C, mmol/L | 2.4±0.6 | 2.3±0.7 | 0.045 |
| eGFR, mL/min/1.73 m2 | 95.6±22.1 | 90.1±21.8 | 0.038 |
| MNSI-PE score | 1.3±0.6 | 3.5±0.7 | <0.001 |
| MNSI-Q score | 1.6±1.7 | 2.8±2.2 | <0.001 |
| MNSI-Q ≥4 | 19 (10.5) | 36 (31.9) | <0.001 |
| 10-g Monofilament <7 | 4 (2.2) | 15 (13.4) | <0.001 |
| Feet ESC, μS | 61.7±14.9 | 55.6±16.1 | 0.001 |
| ESC <56 μS | 56 (30.9) | 48 (42.5) | 0.046 |
| Any clonal hematopoiesis | 36 (19.9) | 10 (8.8) | 0.013 |
| DNMT3A | 15 (8.3) | 3 (2.7) | 0.077 |
| TET2 | 3 (1.7) | 3 (2.7) | 0.679 |
| ASXL1 | 5 (2.8) | 1 (0.9) | 0.412 |
Values are expressed as mean±standard deviation or number (%).
DPN, diabetic peripheral neuropathy; BMI, body mass index; SBP, systolic blood pressure; DBP, diastolic blood pressure; FPG, fasting plasma glucose; HbA1c, hemoglobin A1c; HOMA-IR, homeostatic model assessment for insulin resistance; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; eGFR, estimated glomerular filtration ratio; MNSI-PE, Michigan Neuropathy Screening Instrument-physical examination; MNSI-Q, Michigan Neuropathy Screening Instrument-questionnaire; ESC, electrochemical skin conductance; DNMT3A, DNA methyltransferase 3A; TET2, Tet methylcytosine dioxygenase 2; ASXL1, ASXL transcription regulator.
Table 2.
OR, odds ratio: CI, confidence interval; BMI, body mass index; DM, diabetes mellitus; HbA1c, hemoglobin A1c; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; eGFR, estimated glomerular filtration rate; DNMT3A, DNA methyltransferase 3A; TET2, Tet methylcytosine dioxygenase 2.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download