Abstract
The agenesis of the gonads and adrenal gland in revealed by knockout mouse studies strongly suggested a crucial role for Nr5a1 (SF-1 or Ad4BP) in organ development. In relation to these striking phenotypes, NR5A1/Nr5a1 has the potential to reprogram cells to steroidogenic cells, endow pluripotency, and regulate cell proliferation. However, due to limited knowledge regarding NR5A1 target genes, the mechanism by which NR5A1/Nr5a1 regulates these fundamental processes has remained unknown. Recently, newly-established technologies have enabled the identification of NR5A1 target genes related to multiple metabolic processes, as well as the aforementioned biological processes. Considering that active cellular processes are expected to be accompanied by active metabolism, NR5A1 may act as a key factor for processes such as cell differentiation, proliferation, and survival by coordinating these processes with cellular metabolism. A complete and definite picture of the cellular processes coordinated by NR5A1/Nr5a1 could be depicted by accumulating evidence of the potential target genes through whole genome studies.
NR5A1, also known as steroidogenic factor-1 (SF-1) or adrenal 4-binding protein (Ad4BP), was initially identified as a steroidogenic cell-specific transcription factor regulating the transcription of steroidogenic genes such as cholesterol side chain cleavage enzyme P450 (CYP11A1) gene and steroid 11β hydroxylase P450 (CYP11B1) gene (Fig. 1) [1–3]. Reporter gene assays performed in vitro with cultured cells revealed the involvement of NR5A1 in the regulation of nearly all steroidogenic genes [4,5]. Additional in vivo reporter gene assays performed with transgenic mice [6] and disruption studies of the NR5A1-binding site [7] have confirmed the role of NR5A1 in Cyp11a1 gene transcription. Based on these findings, NR5A1/Nr5a1 has been widely accepted as a key factor for steroidogenic gene transcription.
Another function of Nr5a1 was unveiled through gene knockout (KO) studies. Luo et al. [8] showed the striking phenotypes of Nr5a1 KO mice; the gonads (testis and ovary) and adrenal gland disappeared from the KO mouse fetuses around the time when these organs began to develop. This phenotype was subsequently confirmed by other laboratories [9,10]. In addition to the steroidogenic organs, the functions and structures of the non-steroidogenic pituitary, ventromedial hypothalamic nucleus, and spleen, where Nr5a1 is expressed, were affected in the KO mice [11–14]. Since the phenotypes of the non-steroidogenic organs were less severe than the steroidogenic organs, the functions of NR5A1/Nr5a1 seemed to be different between the two types of the organs. Indeed, many studies have unveiled the functions of NR5A1/Nr5a1 specific for the pituitary [15] and ventromedial hypothalamic nucleus [16–18]. Although the phenotypes differed among the organs, tissues, and cells, these KO mouse studies clearly indicated that Nr5a1 plays fundamental roles in the differentiation, proliferation, and survival of Nr5a1-expressing cells.
The striking phenotypes of the conventional KO mouse led to the anticipation of the unveiled potential of Nr5a1. However, because endocrine regulation in the conventional KO mice was largely disturbed due to the affected gonads, adrenal gland, and pituitary, confirming the observed phenotypes in cell type-specific KO mice was essential. Mouse lines were established using several Cre recombinase (Cre) driver lines [15,18–23]. In this review, we will discuss three conditional KOs from the perspective of the contribution of Nr5a1 to cell differentiation, proliferation, and survival.
Buaas et al. [21] generated a conditional KO mouse using a Cyp11a1-Cre line. Because of the steroidogenic cell-specific expression of Cyp11a1, the Cre line disrupted Nr5a1 only in the steroidogenic cells of the gonad and adrenal cortex. The function and morphology of steroidogenic Leydig cells were affected, and the number of the cells were decreased. However, fetal Leydig cells did not disappear from the KO testes (Fig. 2). In contrast, in a similar study conducted by Shima et al. [22], in which a fetal Leydig-specific enhancer of Nr5a1 was used to drive Cre expression, fetal Leydig cells disappeared from the KO mice. Nr5a1 expression has been believed to increase from the early stage of Leydig cell differentiation [24], and thereafter, in differentiated Leydig cells, NR5A1 activates Cyp11a1 expression. Considering the time lag between Nr5a1 and Cyp11a1 expression, the former conditional KO mice most likely lost Nr5a1 expression after Leydig cell differentiation, while the latter lost it before completion of the differentiation. Thus, conclusively, Nr5a1 is essential for the differentiation of cells but may not be essential for their survival after differentiation.
Anamthathmakula et al. [23] generated Sertoli cell-specific Nr5a1 KO mice using an anti-Müllerian hormone (Amh)-Cre driver line. Amh is known as a marker of Sertoli cells in fetal testes, and Amh expression is regulated by NR5A1 together with other transcription factors [25–28]. Therefore, Nr5a1 was thought to be disrupted soon after the differentiation of Sertoli cells. This timing of Nr5a1 disruption by Amh-Cre in Sertoli cells is likely to correspond to that of Nr5a1 disruption by Cyp11a1-Cre in Leydig cells. Regardless of the similar timing, the number of Sertoli cells largely decreased due to decreased proliferation and increased apoptosis, suggesting that Nr5a1 is required for the proliferation and survival of Sertoli cells. Considering these conditional KO phenotypes, Nr5a1 is likely to play roles in cell differentiation, proliferation, and survival, although the contribution of the gene to these biological processes varies among cell types.
Based on the KO phenotypes, Nr5a1 was speculated to have the potential to differentiate pluripotent cells into gonadal and adrenocortical cells. This potentiality was first discovered by Crawford et al. [29] in 1997 by successfully establishing steroidogenic cells from murine embryonic stem cells through stable Nr5a1 expression alone. Similar results were obtained by Gondo et al. [30] and Yazawa et al. [31] using bone marrow cells and mesenchymal stem cells, respectively. Although these studies indicate the potential for Nr5a1 to contribute to the differentiation of pluripotent stem cells into steroidogenic cells, Nr5a1 alone appeared to be insufficient to specify steroidogenic cells.
Studies of developing gonads have accumulated evidence that sets of transcription factors are required for the differentiation of Sertoli and Leydig cells [32]. Based on these observations, direct reprograming of Sertoli cells from mouse fibroblasts was achieved by Buganim et al. [33]. In this study, the complete reprograming process was divided into three steps. The first step, cell proliferation and the mesenchymal–epithelial transition, was promoted by Nr5a1, Wilms tumor 1 (Wt1), and doublesex and mab-3 related transcription factor 1 (Dmrt 1); the second step, cell aggregation, was promoted by Nr5a1, Wt-1, and SRY-box transcription factor 9 (Sox9); and the last step, conversion to Sertoli cells, was promoted by Nr5a1, Wt-1, Dmrt1, GATA binding protein 4 (Gata4), and Sox9. Notably, Nr5a1 is required for all three steps. By applying a similar experimental design, Liang et al. [34] revealed that two genes, NR5A1 and GATA4, are sufficient for the direct reprograming of human fibroblasts to Sertoli cells. Moreover, under the same concept, Yang et al. [35] successfully reprogramed mouse fibroblasts directly to Leydig cells using Nr5a1, Gata4, and Dmrt1. These studies have shown that Nr5a1/Nr5A1 has the potential to promote whole reprograming steps and, in combination with other transcription factors, to specify the cell types to differentiate.
The aforementioned studies have indicated the indispensable role of Nr5a1 during cell differentiation. NR5A2 (liver receptor homolog-1 [LRH1]), another member of the NR5A subfamily, recognizes the same nucleotide sequences as NR5A1 [2,36]. Therefore, both these NR5A family members were believed to potentially regulate the same sets of target genes. Although their cellular expression is different, the patterns of expression partially overlap. In fact, both NR5A1 and NR5A2 are expressed in the ovary to regulate Cyp11a1 genes [37].
Gu et al. [38] found that Nr5a2 is expressed in the inner cell mass and epiblast of early-stage mouse embryos. Using gene-disrupted mice, they revealed that Nr5a2 in those cells tightly regulates the expression of octamer-binding transcription factor 4 (Oct4), one of the genes essential for reprograming of differentiated cells to pluripotent stem cells. Consistent with this finding, Heng et al. [39] demonstrated that Oct4 can be replaced by Nr5a2 in the reprograming of murine somatic cells to pluripotent cells. Interestingly, both Nr5a1 and Nr5a2 have the potential to reprogram and establish pluripotent cells [38–40]. Given that these two factors share common binding sequences, their common activities were expected to be realized by regulating the same sets of target genes. Two chromatin immunoprecipitation (ChIP)-sequence studies were conducted to identify the target genes of the two factors: one with Nr5a2-expressing mouse fibroblasts [39] and the other with the NR5A1-expressing human embryonic stem cell line (Table 1) [40]. The results obtained from the mouse cells revealed that NR5A2 together with SOX2 and Kruppel like factor 4 (KLF4) regulates genes pivotal for the maintenance of the identity of embryonic stem cells, such as POU domain, class 5, transcription factor 5 (Pou5f1), homeobox transcription factor Nanog (Nanog), T-box transcription factor 3 (Tbx3), and Klf2, while those from the human cells revealed that NR5A1 accumulates at the developmental pluripotency associated 2 (DPPA2) and DPPA4 genes that are known to be key reprograming factors in mice.
Based on the findings of the studies described in this section, NR5A1/Nr5a1 can be inferred to be fundamental for multiple steps of cell differentiation, such as the establishment and reprograming of pluripotent cells and the differentiation of cells to gonadal somatic cells, including steroidogenic cells. Given that future studies would focus on unveiling the mechanism by which NR5A1 regulates these steps of cellular differentiation, identifying NR5A1 target genes is essential to delineate the mechanism.
As describe above, KO phenotypes strongly suggest a role for Nr5a1 in the regulation of cell proliferation. A possible correlation between the amount of Nr5a1 and cell proliferation was suggested for the first time by Bland et al. [41]. They reported that the adrenal glands of Nr5a1 heterozygous mice were smaller than those of wild-type mice [41]. Consistent with this finding, Beuschlein et al. [42] reported that compensatory adrenal growth after unilateral adrenalectomy was strongly affected in heterozygous mice. Conversely, forced Nr5a1 expression resulted in enlargement of the fetal adrenal cortex [43], possibly caused in part by enhanced proliferation [44]. This cell proliferation promotion activity was reproduced with cultured cells [44] and inhibited by inverse agonists of NR5A1 [45].
Along with these studies, the following studies have advanced our understanding of the mechanism by which NR5A1 regulates cell proliferation. Doghman et al. [44] raised the possibility that Nr5a1 regulates cell proliferation by activating FATE1 (a cancer testis antigen) expression. Ishimaru et al. [46] demonstrated that forced expression of Nr5a1 induced cyclin D1 expression, whereby promoting cell proliferation within the chick embryonic gonad. Similarly, Syu et al. [47] demonstrated that proliferation of Y-1 cells was promoted via Nr5a1-activated cyclin E1 expression. In a study investigating cell cycle regulation, Lewis et al. [48] demonstrated that the transcriptional activity of NR5A1 is regulated through phosphorylation by cyclin dependent kinase 7 (CDK7). CDK7, a cyclin-dependent kinase, forms a trimeric complex with cyclin H and Mat1 to act as a CDK-activating kinase complex; this complex is a component of transcription factor IIH (TFIIH) and thus associated with the regulation of basal transcription [49]. Taken together, CDK7 might be assumed to serve as a direct link between cell cycle progression through activation of other CDKs and transcription through NR5A1 phosphorylation. Moreover, CDK7 could couple NR5A1-driven transcriptional activation with activation of the basal transcriptional machinery.
Regarding regulation of cell proliferation by Nr5a1, interesting observations were reported by Lai et al. [50] and Wang et al. [51]. A series of studies reported that NR5A1 was localized at the centrosome. Dissociation of NR5A1 from the centrosome promoted DNA-dependent protein kinase (DNA-PK) recruitment and thereby activated CDK2, whose activity is required for the duplication of DNA and the centrosome. When Nr5a1 is knocked down, CDK2 is aberrantly activated by DNA-PK, and thus, the centrosome is over-duplicated. Consequently, cell proliferation is disordered. Although the studies described in this section are still fragmented rather than tightly connected, evidence supporting the role of Nr5a1 in cell cycle regulation has accumulated gradually from a wide variety of sources.
Early studies of Nr5a1 predominantly focused on steroidogenic genes. Thereafter, studies were conducted to identify target genes in nonsteroidogenic cells, such as Sertoli cells and pituitary gonadotrophs. Consequently, in addition to steroidogenic genes, several nonsteroidogenic gene targets were identified. However, it seemed unlikely that NR5A1, merely thought to regulate genes already identified, was responsible for a wide range of biological processes, such as differentiation and reprograming to particular cell types, cell proliferation, or survival. To comprehensively understand the actions of Nr5a1, genome-wide studies with DNA arrays and deep sequencing were conducted (Table 1) [40,52–58]. In fact, these genome-wide studies identified many novel target genes of NR5A1, and eventually unveiled novel roles of NR5A1 in biological, physiological, and pathological processes.
Human cohort studies have shown that strong NR5A1 expression in adrenocortical carcinoma correlates with a poor clinical outcome [59]. However, it remained to be clarified which genes, under the control of the highly-expressed NR5A1, were responsible for the poor prognosis. Through ChIP-seq, Ruggiero et al. [56] identified guanine nucleotide exchange factor 2 (VAV2) as one of the NR5A1 target genes in H295R cells (a human adrenocortical carcinoma cell line). VAV2, a member of the VAV family, has been characterized as a guanine exchange factor that activates the Rho/Rac family of GTPases, and thus promotes cellular remodeling and invasion [60]. Ruggiero et al. [56] clearly demonstrated that NR5A1-induced VAV2 activates the small GTPases cell division cycle 42 (CDC42) and Rac family small GTPase 1 (RAC1), and consequently promotes cytoskeleton remodeling and cell invasion. This result could provide the rationale behind highly-expressed NR5A1 causing poor clinical outcomes in adrenocortical carcinoma.
By using Y-1 adrenocortical and testicular Leydig cells, the role of Nr5a1 in the regulation of energy metabolism-related genes was identified through ChIP-seq. Baba et al. [55] demonstrated that nearly all glycolytic genes are regulated by NR5A1 (Fig. 3). Indeed, Nr5a1 knockdown resulted in a decrease in glucose consumption as well as in glycolytic gene expression. Along with the tricarboxylic acid cycle and oxidative phosphorylation, glycolysis is the main pathway for supplying the energized molecule, adenosine triphosphate (ATP). As expected, intracellular ATP concentration decreased with Nr5a1 knockdown. Another energized molecule, nicotinamide-adenine dinucleotide phosphate (NADPH, reduce form), is required for the synthesis of various biomolecules. Steroidogenic reactions mediated by cytochrome P450s consume NADPH [61]. This energized molecule is synthesized by multiple pathways and enzymes, such as the pentose phosphate pathway, malic enzymes, and methylenetetrahydrofolate dehydrogenases. Among these, genes encoding malic enzyme 1 and methylenetetrahydrofolate dehydrogenase 2 were identified as Nr5a1 targets [57]. NADPH concentration also decreased with Nr5a1 knockdown.
Moreover, many cholesterogenic genes were shown to be NR5A1 target genes by Baba et al. [58]. Concordantly, cholesterogenic activity was decreased by Nr5a1 knockdown. Cholesterol is utilized as a starting material for multiple steroidogenic reactions, wherein the first reaction is mediated by the cholesterol side-chain cleavage P450 localized in mitochondria. Therefore, cholesterol must be transported into mitochondria. Hypoxia up-regulated mitochondrial movement regulator (HUMMR)/mitochondria-localized glutamic acid-rich protein (MGARP) promotes cholesterol transfer to the mitochondrial outer membrane [62], and interestingly, the Hummr/Mgarp-encoding gene is an NR5A1 target together with cholesterogenic genes. Expectedly, the amount of cholesterol in mitochondria decreased with Nr5a1 knockdown [58].
Many studies have demonstrated that cell type-specific transcription factors are specialized for the regulation of cell-specific genes. However, genome-wide studies have revealed that NR5A1, regardless of its nature as a cell type-specific transcription factor, regulates housekeeping processes such as glycolysis, NADPH synthesis, and cholesterogenesis. As shown in Fig. 4, ATP and NADPH are essential cofactors for cholesterogenesis and steroidogenesis. Moreover, cholesterol is the starting material for steroidogenesis. Therefore, efficient steroidogenesis cannot be achieved unless these materials are supplied simultaneously and efficiently. Accordingly, NR5A1/Nr5a1 is assumed to be responsible for achieving efficient steroidogenesis through the orchestration of multiple types of metabolisms.
Similarly, the role of NR5A1/Nr5a1 in cell differentiation, proliferation, survival, and invasion could be understood in correlation with its ability to regulate metabolisms. When cells proliferate, for instance, they produce various components and assemble them into cellular structures by consuming large amounts of energized molecules. By coordinating between the metabolisms and the above cellular processes, NR5A1/Nr5a1 may act as a cell-specific factor.
Although many target genes have been identified through ChIP-seq studies, we may have not yet comprehensively unveiled the biological significance of NR5A1/Nr5a1. Depicting the whole view by assembling jigsaw pieces could eventually provide a rationale for the disappearance of the gonads and adrenal glands from the KO mice, and at the same time, could exemplify the integration of multiple cellular processes by a single transcription factor.
ACKNOWLEDGMENTS
The authors are supported by grants JSPS KAKENHI Grant Number JP20H03436 (Ken-ichirou Morohashi), JP17H06427 (Ken-ichirou Morohashi, Takashi Baba), and JP20K08863 (Takashi Baba); by AMED under Grant Number JP20gk0210019 (Ken-ichirou Morohashi).
REFERENCES
1. Lala DS, Rice DA, Parker KL. Steroidogenic factor I, a key regulator of steroidogenic enzyme expression, is the mouse homolog of fushi tarazu-factor I. Mol Endocrinol. 1992; 6:1249–58.

2. Morohashi K, Honda S, Inomata Y, Handa H, Omura T. A common trans-acting factor, Ad4-binding protein, to the promoters of steroidogenic P-450s. J Biol Chem. 1992; 267:17913–9.

3. Honda S, Morohashi K, Nomura M, Takeya H, Kitajima M, Omura T. Ad4BP regulating steroidogenic P-450 gene is a member of steroid hormone receptor superfamily. J Biol Chem. 1993; 268:7494–502.

4. Morohashi Ki. Gonadal and extragonadal functions of Ad4BP/SF-1: developmental aspects. Trends Endocrinol Metab. 1999; 10:169–73.

5. Parker KL, Schimmer BP. Steroidogenic factor 1: a key determinant of endocrine development and function. Endocr Rev. 1997; 18:361–77.

6. Hu MC, Hsu NC, Pai CI, Wang CK, Chung BC. Functions of the upstream and proximal steroidogenic factor 1 (SF-1)-binding sites in the CYP11A1 promoter in basal transcription and hormonal response. Mol Endocrinol. 2001; 15:812–8.

7. Shih MC, Hsu NC, Huang CC, Wu TS, Lai PY, Chung BC. Mutation of mouse Cyp11a1 promoter caused tissue-specific reduction of gene expression and blunted stress response without affecting reproduction. Mol Endocrinol. 2008; 22:915–23.
8. Luo X, Ikeda Y, Parker KL. A cell-specific nuclear receptor is essential for adrenal and gonadal development and sexual differentiation. Cell. 1994; 77:481–90.

9. Sadovsky Y, Crawford PA, Woodson KG, Polish JA, Clements MA, Tourtellotte LM, et al. Mice deficient in the orphan receptor steroidogenic factor 1 lack adrenal glands and gonads but express P450 side-chain-cleavage enzyme in the placenta and have normal embryonic serum levels of corticosteroids. Proc Natl Acad Sci U S A. 1995; 92:10939–43.

10. Morohashi KI, Omura T. Ad4BP/SF-1, a transcription factor essential for the transcription of steroidogenic cytochrome P450 genes and for the establishment of the reproductive function. FASEB J. 1996; 10:1569–77.

11. Ingraham HA, Lala DS, Ikeda Y, Luo X, Shen WH, Nachtigal MW, et al. The nuclear receptor steroidogenic factor 1 acts at multiple levels of the reproductive axis. Genes Dev. 1994; 8:2302–12.

12. Shinoda K, Lei H, Yoshii H, Nomura M, Nagano M, Shiba H, et al. Developmental defects of the ventromedial hypothalamic nucleus and pituitary gonadotroph in the Ftz-F1 disrupted mice. Dev Dyn. 1995; 204:22–9.

13. Ikeda Y, Luo X, Abbud R, Nilson JH, Parker KL. The nuclear receptor steroidogenic factor 1 is essential for the formation of the ventromedial hypothalamic nucleus. Mol Endocrinol. 1995; 9:478–86.

14. Morohashi K, Tsuboi-Asai H, Matsushita S, Suda M, Nakashima M, Sasano H, et al. Structural and functional abnormalities in the spleen of an mFtz-F1 gene-disrupted mouse. Blood. 1999; 93:1586–94.
15. Zhao L, Bakke M, Parker KL. Pituitary-specific knockout of steroidogenic factor 1. Mol Cell Endocrinol. 2001; 185:27–32.

16. Segal JP, Stallings NR, Lee CE, Zhao L, Socci N, Viale A, et al. Use of laser-capture microdissection for the identification of marker genes for the ventromedial hypothalamic nucleus. J Neurosci. 2005; 25:4181–8.

17. Tran PV, Akana SF, Malkovska I, Dallman MF, Parada LF, Ingraham HA. Diminished hypothalamic bdnf expression and impaired VMH function are associated with reduced SF-1 gene dosage. J Comp Neurol. 2006; 498:637–48.
18. Kim KW, Zhao L, Parker KL. Central nervous system-specific knockout of steroidogenic factor 1. Mol Cell Endocrinol. 2009; 300:132–6.

19. Jeyasuria P, Ikeda Y, Jamin SP, Zhao L, De Rooij DG, Themmen AP, et al. Cell-specific knockout of steroidogenic factor 1 reveals its essential roles in gonadal function. Mol Endocrinol. 2004; 18:1610–9.

20. Pelusi C, Ikeda Y, Zubair M, Parker KL. Impaired follicle development and infertility in female mice lacking steroidogenic factor 1 in ovarian granulosa cells. Biol Reprod. 2008; 79:1074–83.
21. Buaas FW, Gardiner JR, Clayton S, Val P, Swain A. In vivo evidence for the crucial role of SF1 in steroid-producing cells of the testis, ovary and adrenal gland. Development. 2012; 139:4561–70.

22. Shima Y, Miyabayashi K, Sato T, Suyama M, Ohkawa Y, Doi M, et al. Fetal Leydig cells dedifferentiate and serve as adult Leydig stem cells. Development. 2018; 145:dev169136.

23. Anamthathmakula P, Miryala CSJ, Moreci RS, Kyathanahalli C, Hassan SS, Condon JC, et al. Steroidogenic factor 1 (Nr5a1) is required for sertoli cell survival post sex determination. Sci Rep. 2019; 9:4452.

24. Miyabayashi K, Katoh-Fukui Y, Ogawa H, Baba T, Shima Y, Sugiyama N, et al. Aristaless related homeobox gene, Arx, is implicated in mouse fetal Leydig cell differentiation possibly through expressing in the progenitor cells. PLoS One. 2013; 8:e68050.

25. De Santa Barbara P, Bonneaud N, Boizet B, Desclozeaux M, Moniot B, Sudbeck P, et al. Direct interaction of SRY-related protein SOX9 and steroidogenic factor 1 regulates transcription of the human anti-Müllerian hormone gene. Mol Cell Biol. 1998; 18:6653–65.

26. Tremblay JJ, Viger RS. Transcription factor GATA-4 enhances Müllerian inhibiting substance gene transcription through a direct interaction with the nuclear receptor SF-1. Mol Endocrinol. 1999; 13:1388–401.
27. Watanabe K, Clarke TR, Lane AH, Wang X, Donahoe PK. Endogenous expression of Müllerian inhibiting substance in early postnatal rat sertoli cells requires multiple steroidogenic factor-1 and GATA-4-binding sites. Proc Natl Acad Sci U S A. 2000; 97:1624–9.

28. Lasala C, Carre-Eusebe D, Picard JY, Rey R. Subcellular and molecular mechanisms regulating anti-Müllerian hormone gene expression in mammalian and nonmammalian species. DNA Cell Biol. 2004; 23:572–85.

29. Crawford PA, Sadovsky Y, Milbrandt J. Nuclear receptor steroidogenic factor 1 directs embryonic stem cells toward the steroidogenic lineage. Mol Cell Biol. 1997; 17:3997–4006.

30. Gondo S, Yanase T, Okabe T, Tanaka T, Morinaga H, Nomura M, et al. SF-1/Ad4BP transforms primary long-term cultured bone marrow cells into ACTH-responsive steroidogenic cells. Genes Cells. 2004; 9:1239–47.

31. Yazawa T, Mizutani T, Yamada K, Kawata H, Sekiguchi T, Yoshino M, et al. Differentiation of adult stem cells derived from bone marrow stroma into Leydig or adrenocortical cells. Endocrinology. 2006; 147:4104–11.

32. Rotgers E, Jorgensen A, Yao HH. At the crossroads of fate-somatic cell lineage specification in the fetal gonad. Endocr Rev. 2018; 39:739–59.

33. Buganim Y, Itskovich E, Hu YC, Cheng AW, Ganz K, Sarkar S, et al. Direct reprogramming of fibroblasts into embryonic Sertoli-like cells by defined factors. Cell Stem Cell. 2012; 11:373–86.

34. Liang J, Wang N, He J, Du J, Guo Y, Li L, et al. Induction of Sertoli-like cells from human fibroblasts by NR5A1 and GATA4. Elife. 2019; 8:e48767.

35. Yang Y, Li Z, Wu X, Chen H, Xu W, Xiang Q, et al. Direct reprogramming of mouse fibroblasts toward Leydig-like cells by defined factors. Stem Cell Reports. 2017; 8:39–53.

36. Fayard E, Auwerx J, Schoonjans K. LRH-1: an orphan nuclear receptor involved in development, metabolism and steroidogenesis. Trends Cell Biol. 2004; 14:250–60.

37. Saxena D, Escamilla-Hernandez R, Little-Ihrig L, Zeleznik AJ. Liver receptor homolog-1 and steroidogenic factor-1 have similar actions on rat granulosa cell steroidogenesis. Endocrinology. 2007; 148:726–34.

38. Gu P, Goodwin B, Chung AC, Xu X, Wheeler DA, Price RR, et al. Orphan nuclear receptor LRH-1 is required to maintain Oct4 expression at the epiblast stage of embryonic development. Mol Cell Biol. 2005; 25:3492–505.

39. Heng JC, Feng B, Han J, Jiang J, Kraus P, Ng JH, et al. The nuclear receptor Nr5a2 can replace Oct4 in the reprogramming of murine somatic cells to pluripotent cells. Cell Stem Cell. 2010; 6:167–74.

40. Yamauchi K, Ikeda T, Hosokawa M, Nakatsuji N, Kawase E, Chuma S, et al. Overexpression of nuclear receptor 5a1 induces and maintains an intermediate state of conversion between primed and naive pluripotency. Stem Cell Reports. 2020; 14:506–19.

41. Bland ML, Jamieson CA, Akana SF, Bornstein SR, Eisenhofer G, Dallman MF, et al. Haploinsufficiency of steroidogenic factor-1 in mice disrupts adrenal development leading to an impaired stress response. Proc Natl Acad Sci U S A. 2000; 97:14488–93.
42. Beuschlein F, Mutch C, Bavers DL, Ulrich-Lai YM, Engeland WC, Keegan C, et al. Steroidogenic factor-1 is essential for compensatory adrenal growth following unilateral adrenalectomy. Endocrinology. 2002; 143:3122–35.

43. Zubair M, Oka S, Parker KL, Morohashi K. Transgenic expression of Ad4BP/SF-1 in fetal adrenal progenitor cells leads to ectopic adrenal formation. Mol Endocrinol. 2009; 23:1657–67.
44. Doghman M, Karpova T, Rodrigues GA, Arhatte M, De Moura J, Cavalli LR, et al. Increased steroidogenic factor-1 dosage triggers adrenocortical cell proliferation and cancer. Mol Endocrinol. 2007; 21:2968–87.

45. Doghman M, Cazareth J, Douguet D, Madoux F, Hodder P, Lalli E. Inhibition of adrenocortical carcinoma cell proliferation by steroidogenic factor-1 inverse agonists. J Clin Endocrinol Metab. 2009; 94:2178–83.

46. Ishimaru Y, Komatsu T, Kasahara M, Katoh-Fukui Y, Ogawa H, Toyama Y, et al. Mechanism of asymmetric ovarian development in chick embryos. Development. 2008; 135:677–85.

47. Syu JS, Baba T, Huang JY, Ogawa H, Hsieh CH, Hu JX, et al. Lysosomal activity maintains glycolysis and cyclin E1 expression by mediating Ad4BP/SF-1 stability for proper steroidogenic cell growth. Sci Rep. 2017; 7:240.

48. Lewis AE, Rusten M, Hoivik EA, Vikse EL, Hansson ML, Wallberg AE, et al. Phosphorylation of steroidogenic factor 1 is mediated by cyclin-dependent kinase 7. Mol Endocrinol. 2008; 22:91–104.

49. Fisher RP. Secrets of a double agent: CDK7 in cell-cycle control and transcription. J Cell Sci. 2005; 118:5171–80.

50. Lai PY, Wang CY, Chen WY, Kao YH, Tsai HM, Tachibana T, et al. Steroidogenic factor 1 (NR5A1) resides in centrosomes and maintains genomic stability by controlling centrosome homeostasis. Cell Death Differ. 2011; 18:1836–44.

51. Wang CY, Kao YH, Lai PY, Chen WY, Chung BC. Steroidogenic factor 1 (NR5A1) maintains centrosome homeostasis in steroidogenic cells by restricting centrosomal DNA-dependent protein kinase activation. Mol Cell Biol. 2013; 33:476–84.

52. Ferraz-de-Souza B, Lin L, Shah S, Jina N, Hubank M, Dattani MT, et al. ChIP-on-chip analysis reveals angiopoietin 2 (Ang2, ANGPT2) as a novel target of steroidogenic factor-1 (SF-1, NR5A1) in the human adrenal gland. FASEB J. 2011; 25:1166–75.

53. Ju Y, Mizutani T, Imamichi Y, Yazawa T, Matsumura T, Kawabe S, et al. Nuclear receptor 5A (NR5A) family regulates 5-aminolevulinic acid synthase 1 (ALAS1) gene expression in steroidogenic cells. Endocrinology. 2012; 153:5522–34.

54. Doghman M, Figueiredo BC, Volante M, Papotti M, Lalli E. Integrative analysis of SF-1 transcription factor dosage impact on genome-wide binding and gene expression regulation. Nucleic Acids Res. 2013; 41:8896–907.

55. Baba T, Otake H, Sato T, Miyabayashi K, Shishido Y, Wang CY, et al. Glycolytic genes are targets of the nuclear receptor Ad4BP/SF-1. Nat Commun. 2014; 5:3634.

56. Ruggiero C, Doghman-Bouguerra M, Sbiera S, Sbiera I, Parsons M, Ragazzon B, et al. Dosage-dependent regulation of VAV2 expression by steroidogenic factor-1 drives adrenocortical carcinoma cell invasion. Sci Signal. 2017; 10:eaal2464.
57. Li B, Baba T, Miyabayashi K, Sato T, Shima Y, Ichinose T, et al. Role of Ad4-binding protein/steroidogenic factor 1 in regulating NADPH production in adrenocortical Y-1 cells. Endocr J. 2017; 64:315–24.

58. Baba T, Otake H, Inoue M, Sato T, Ishihara Y, Moon JY, et al. Ad4BP/SF-1 regulates cholesterol synthesis to boost the production of steroids. Commun Biol. 2018; 1:18.

59. Sbiera S, Schmull S, Assie G, Voelker HU, Kraus L, Beyer M, et al. High diagnostic and prognostic value of steroidogenic factor-1 expression in adrenal tumors. J Clin Endocrinol Metab. 2010; 95:E161–71.

Fig. 1
Genes implicated in steroid hormone synthetic pathways. Pathways for the synthesis of cortisol (glucocorticoids) and aldosterone (mineralocorticoids) in the adrenal cortex, and testosterone (androgen) and 17β-estradiol (estrogen) in the gonads from cholesterol are shown. Genes implicated in the pathways are indicated in the closed boxes.
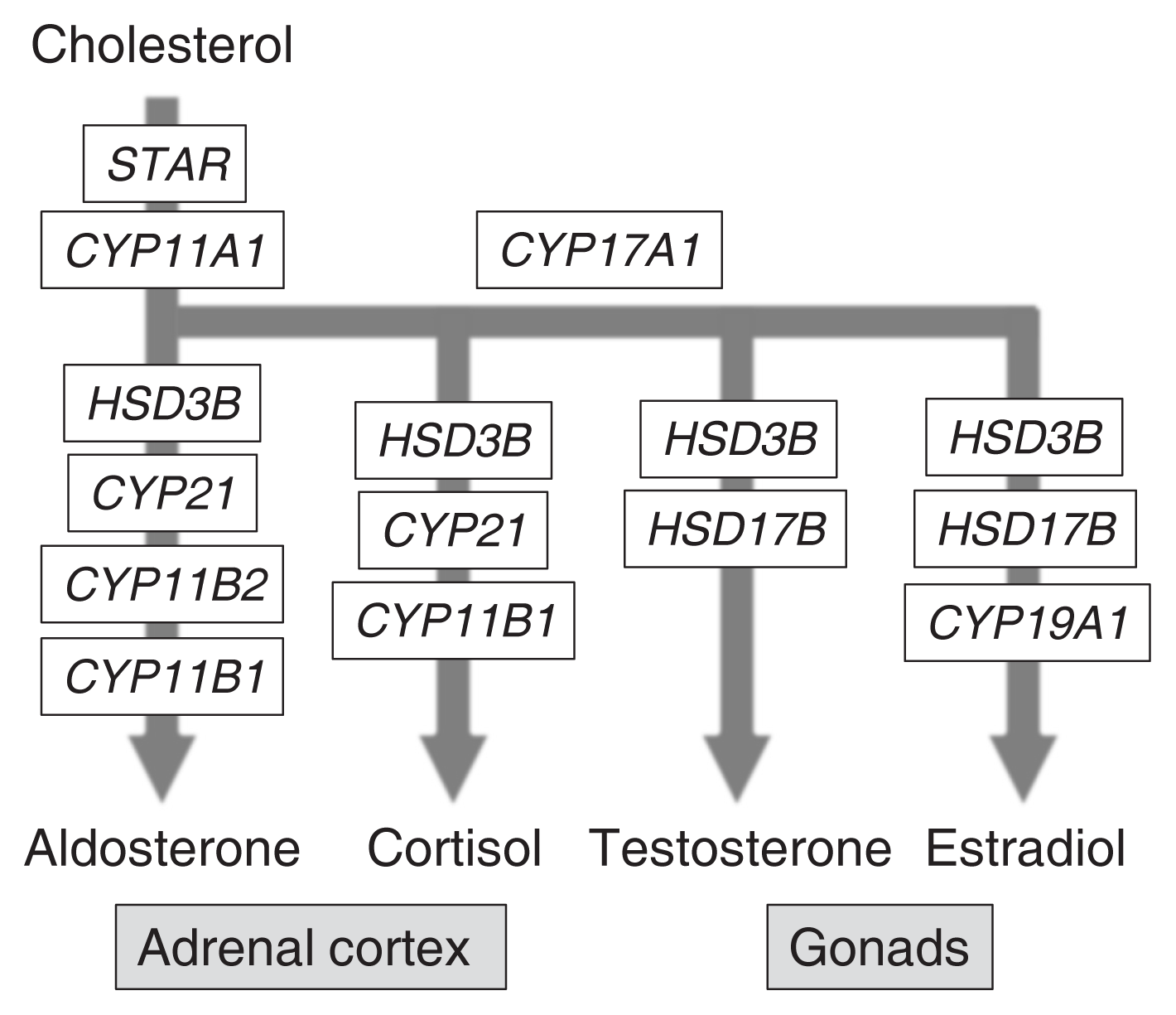
Fig. 2
Effects of cell type-specific Nr5a1 disruption. Nr5a1 expression starts at the progenitor cell stage and reaches a plateau in matured fetal Leydig cells (FLCs) and Sertoli cells. (A) Nr5a1 was disrupted in FLCs or progenitor cells using the Cyp11a1-Cre or FLE-Cre mouse line, respectively. Thus, Nr5a1 expression (indicated by broad arrows) was expected to increase once and then be abolished in the FLCs or progenitor cells (broken arrows). (B) Nr5a1 was disrupted in Sertoli cells using the Amh-Cre mouse line. The expression was expected to increase once and then be abolished after the differentiation of Sertoli cells (indicated by a broad arrow and a broken arrow). The effects of Nr5a1 on the presence of the cell types and the cellular functions in the conditional knockout mice are summarized on the right side.
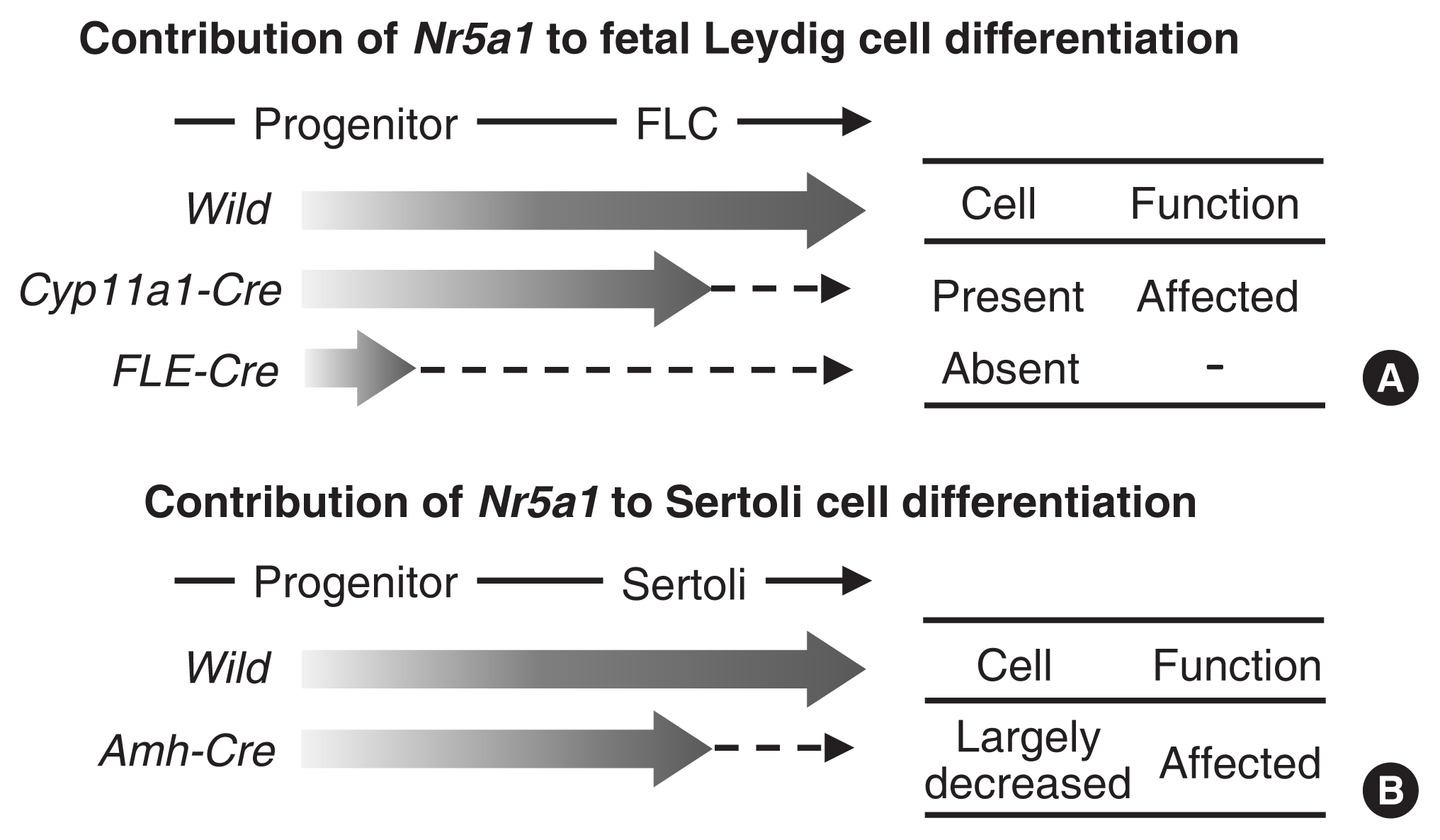
Fig. 3
Cellular metabolisms regulated by Nr5a1. Chromatin immunoprecipitation (ChIP)-seq studies revealed that NR5A1 regulates genes involved in glycolysis (shown by an orange arrow), cholesterogenesis (blue arrow), and steroidogenesis (blue arrow). In addition, NR5A1 regulates genes whose products mediate nicotinamide-adenine dinucleotide phosphate (NADPH) synthesis (orange arrow). Glycolysis and NADPH synthesis pathways produce the energized molecules adenosine triphosphate (ATP) and NADPH, respectively, whereas cholesterogenic and steroidogenic pathways consume these energized molecules. Because pyruvate, the product of glycolysis, can be converted to acetyl-coenzyme A (CoA), the starting material for cholesterogenesis, and cholesterol is used for steroidogenesis, Nr5a1 might thus be considered to coordinate multiple metabolic pathways to achieve efficient steroidogenesis.
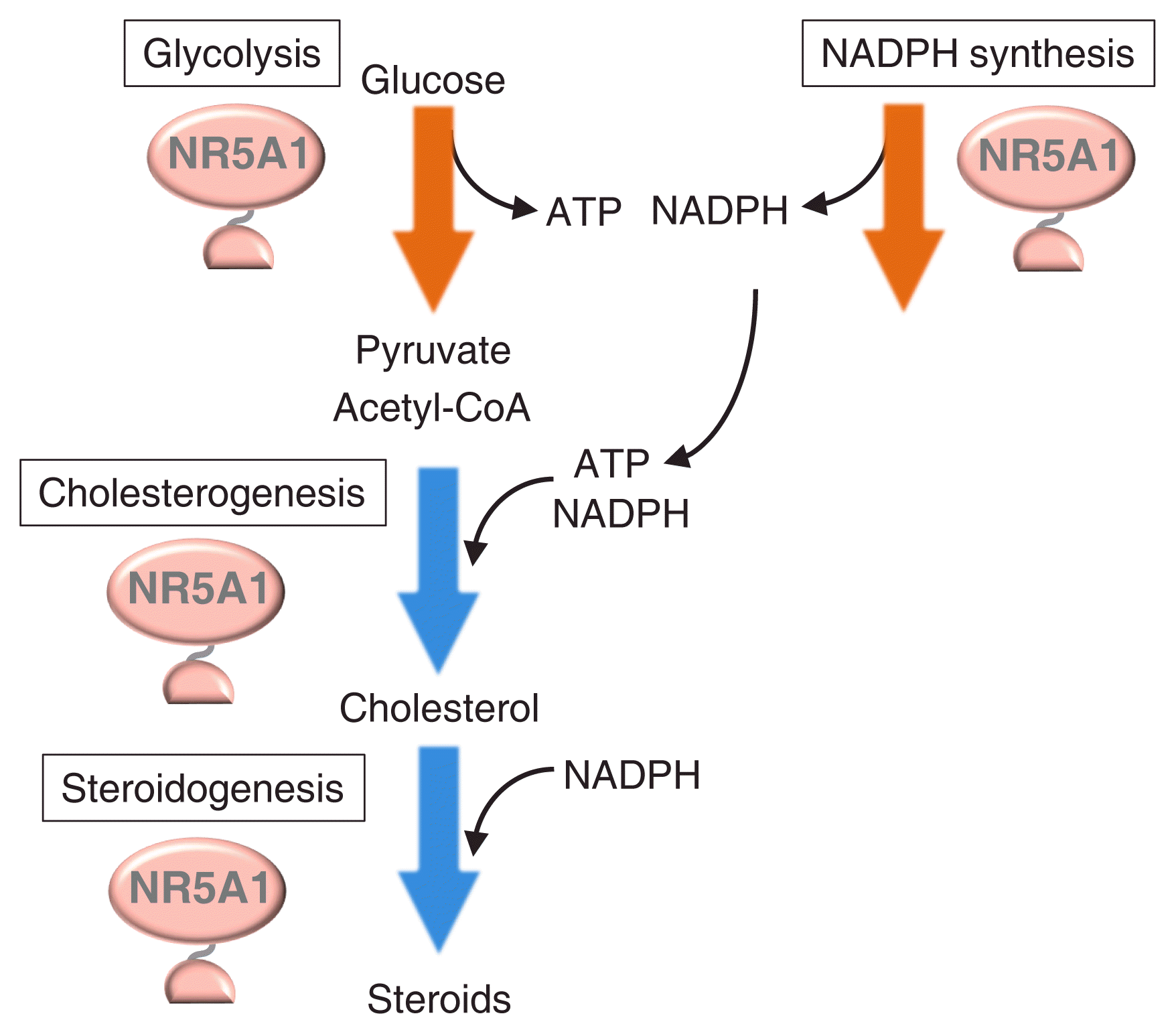
Fig. 4
Possible coordination between cellular activities and metabolisms by Nr5a1. Genes involved in multiple metabolic pathways, such as glycolysis, cholesterogenesis, steroidogenesis, and nicotinamide-adenine dinucleotide phosphate (NADPH) synthesis, are regulated by Nr5a1. Nr5a1 potentially regulates the processes of cell proliferation, differentiation, and survival. Synthesis of DNA, RNA, proteins, and lipids are thought to accompany these cellular activities. The energized molecules, adenosine triphosphate (ATP) and/or NADPH, are required for the production of these cellular components. By supplying the energized molecules, Nr5a1 may coordinate cellular metabolisms and activities. CoA, coenzyme A.
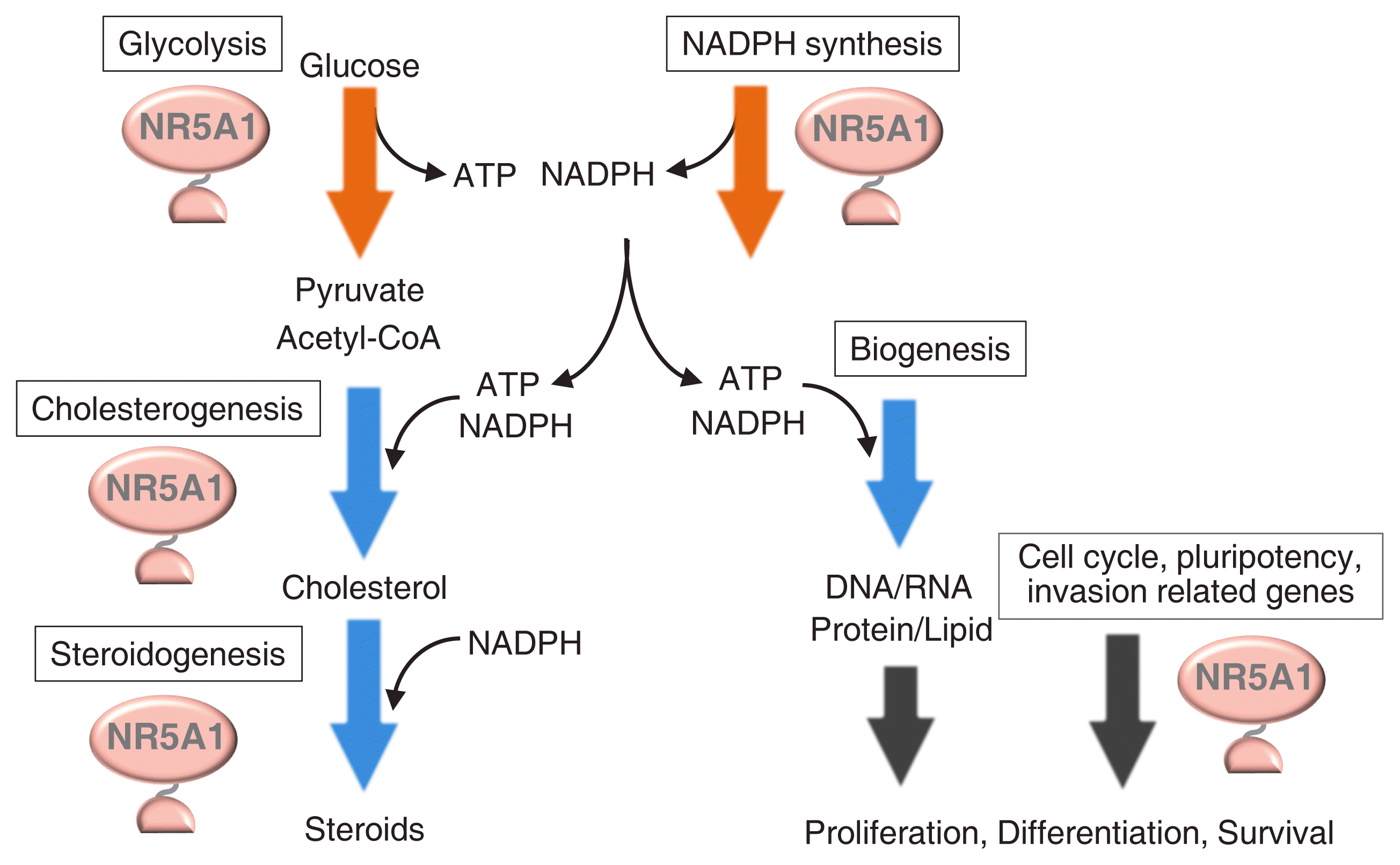
Table 1
Genome Wide Studies to Identify NR5A1 Target Genes
| Study | Method | Cell (species) |
|---|---|---|
| Ferraz-de-Souza et al. (2011) [52] | ChIP-on-chip | H295 (human) |
| Ju et al. (2012) [53] | ChIP-on-chip | Mesenthymal stem cell (mouse) |
| Doghman et al. (2013) [54] | ChIP-seq | H295R (human) |
| Baba et al. (2014) [55] | ChIP-seq | Y-1, Leydig cell (mouse) |
| Ruggiero et al. (2017) [56] | ChIP-seq | H295R (human) |
| Li et al. (2017) [57] | ChIP-seq | Y-1, mouse (mouse) |
| Baba et al. (2018) [58] | ChIP-seq | Y-1, Leydig cell (mouse) |
| Yamauchi et al. (2020) [40] | ChIP-seq | Embryonic stem cell (human) |




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download