INTRODUCTION
Canine distemper virus (CDV) of the genus Morbillivirus in the family Paramyxoviridae is a fatal dog pathogen. CDV infections have been reported in mink, raccoon dogs, tigers, lions, and monkeys (1, 2, 3). CDV (15,690 nucleotides) encodes eight proteins: the nucleocapsid (N), phosphoprotein (P), non-structural (C and V), matrix (M), fusion (F), hemagglutinin (H), and large (L) proteins (4). Of these, the H and F proteins play key roles in penetration into receptor-bearing cells and in induction of an immune response (4, 5). The H protein promotes virus binding to the host cell membrane, and the F protein fuses the virus with the membrane (5).
CDV vaccines (commercial distemper/adeno/parvo/parainfluenza (DAPP) vaccines containing CDV, canine adenovirus type 1 (CAV-1) or canine adenovirus type 2 (CAV-2), canine parvovirus (CPV), and canine parainfluenza virus (CPIV) are routinely used to inoculate South Korean dogs (6). Most CDV vaccine strains are designed against the Lederle, Onderstepoort, and Rockborn strains of the America-1 genotype and were developed in the 1950s (7). Vaccination has dramatically reduced CDV infections in 4–8-week-old dogs. However, since the 1990s, canine distemper infection in vaccinated dogs has been reported in many countries, including China, Mexico, Finland, and Japan (8, 9, 10, 11). Also, the CDV vaccines are not recommended for wild carnivores including zoo animals because of the possibility that attenuation is inadequate (12). CDV infections continue to be reported in South Korean dogs and raccoon dogs because of CDV variation (2, 13). The CDVs circulating in South Korea have been molecularly confirmed to be of the Asia-1 or -2 genotype (2). Therefore, development of new CDV vaccines against these strains, and evaluation of existing CDV vaccines, are required.
The signaling lymphocyte activation molecule (SLAM), CD46, and nectin-4 (14, 15, 16) of Vero cells expressing the dog SLAM gene serve as morbillivirus receptors. The cells have been used to isolate wild CDVs. We previously reported that CD1901, isolated from a naturally infected dog in 2019, grew in Vero/dSLAM cells and induced a cytopathic effect (CPE) such as cell fusion (17). Vero cells expressing viral receptors are useful not only for virus isolation and propagation, but also for elucidating the biological properties of newly isolated CDVs (14). Few reports have described selection of CDV vaccine candidates on passage through several cell lines, including Vero cells expressing viral receptors (7).
Here, we constructed a Vero cell line expressing the dog nectin-4 gene and passaged CD1901 100 times in four types of cells and named it CD1901-100. We investigated the biological and molecular properties of the CD1901-100 strain in terms of biological manifestations and whole-genome sequence data. This information will serve as the basis for the development of inactivated or live CDV vaccine strains.
Go to : 
MATERIALS AND METHODS
Virus and cells
A new Korean CDV was isolated in 2019 from a naturally infected Korean dog and named CD1901 (17). This strain was continuously passaged 100 times in Vero/dSLAM, Vero (ATCC, CCL 81), DF-1 (ATCC, CRL12203), and Vero/dNectin-4 cells, and named CD1901-100. The four cell types were cultured in DMEM supplemented with penicillin and streptomycin, an antimycotic, and 10% (v/v) fetal bovine serum.
Construction of Vero/dNectin-4 cells
We constructed Vero/dNectin4 cells to improve the proliferative capacity of CDV. The dog nectin-4 gene was inserted into a lentiviral expression vector to create pcLV-CMV-dog nectin-4-eGFP-puro, which contains both green fluorescent protein (GFP) and puromycin resistance genes. The lentivirus containing the dog nectin-4 gene was inoculated into Vero cells when the cells attained 30% confluence on a six-well plate. The 2nd passaged cells were named Vero/dNectin-4 cells after confirming dog nectin-4 gene expression via both fluorescence microscopy and polymerase chain reaction (PCR). Fluorescents of Vero/dNectin-4 cells and normal Vero cells were observed with fluorescence microscope (TE2000-U, Nikon Instruments Inc., Tokyo, Japan). DNA was extracted using a kit (Bioneer, Daejeon, Korea) and subjected to PCR employing two primer sets to amplify partial dog nectin-4 genes (Table 1).
Table 1.
Polymerase chain reaction primers used to amplify the dog nectin-4 gene of Vero/dNectin-4 cells
| Name of primer | Oligonucleotide (5'-3') | Size (bp) | Target gene |
|---|---|---|---|
| NecF1 | ATGCCTCTATCCCTGGGAGC | 240 | Nectin-4 |
| NecR1 | CAGGGCTAATTCCCGGGCAG | ||
| NecF2 | GGGCAGGGCCTGACATTGGC | 360 | Nectin-4 |
| NecR2 | ATTGTATGAGGGAGGGGGGTGC |
CD1901 passage
CD1901 was passaged in four types of cells to derive a new CDV with excellent proliferative capacity. The 10-fold diluted viruses were inoculated into the cells and harvested from the last wells showing the CPE. As shown in Table 2, CD1901 was passaged 40 times in Vero/dSLAM cells without any treatment and then again (passages 41 to 60) after 1 min of ultraviolet (UV) light exposure about 60 centimeters away in a biosafety cabinet. Passages 61 to 76 employed DF-1 cells, and passages 77 to 84 used normal Vero cells. Passages 85-95 employed Vero/dSLAM cells and were performed in the presence of 4 mM 5' bromouracil. Passages 96-100 used Vero/dNectin-4 cells.
Cytopathic effects and immunofluorescence assay (IFA)
Vero/dSLAM cells infected with CD1901 and CD1901-100 in six-well plates were microscopically compared 5 days post-inoculation. For the immunofluorescence assay, Vero/dSLAM cells infected with CD1901 and CD1901-100 in six-well plates were fixed in cold acetone, reacted with a mouse monoclonal antibody against CDV (VMRD, Pullman, WA, USA) at 37°C for 1 h, and then stained with fluorescent isothiocyanate-conjugated goat-anti-mouse IgG + IgM antibodies (KPL Laboratories, Gettysburg, PA, USA). After washing with phosphate-buffered saline (pH 7.2), cytoplasmic fluorescents of Vero/dSLAM cells inoculated with CD1901 and CD1901-100 were compared under a 100x fluorescence microscope.
Growth kinetics, titration, and cell tropism
We measured the growth kinetics of CD1901 and CD1901-100. Vero/dSLAM cells grown in 25-cm2 flasks were inoculated with the viruses at 103.0 TCID50/mL and harvested daily for 8 days. The growth kinetics were derived as previously reported (17). The titers were determined using the Reed and Muench method (18) and expressed as the 50% tissue culture infectious doses/mL (TCID50/mL).
CD1901 and CD1901-100 were inoculated into Vero/dSLAM, Vero/hSLAM (ECACC General Collection, London, England), Vero/dNectin-4, Vero, DF-1, A72 (ATCC, CRL-1542), and MDCK (ATCC, CRL34) cells and grown in 25-cm2 flasks. After incubation for 5 days, each flask was frozen and thawed three times. The clarified supernatants were subjected to viral titration to know the proliferative ability of CD1901-100 as described above.
Whole-genome sequencing (WGS) and phylogenetic analysis
RNAs of both CD1901 and CD1901-100 were extracted with the Qiagen Viral RNA Mini Kit, and were whole-genome sequenced by Senigen (Anyang, Korea). Reverse transcription employed Superscript IV reverse transcriptase (ThermoFisher Scientific, Waltham, MA, USA). Barcoded sequencing libraries of both strains were prepared using TruSeq® Nano DNA LT kits (Illumina, California, USA). cDNA was sonicated using a ultrasonicator (Covaris, Massachusetts, USA). The fragmented DNA was subjected to end-repair PCR. Indices were attached to blunt fragments. The barcoded DNA fragments were enriched and cleaned in accordance with Illumina TruSeq Nano DNA sample preparation guide. The amplicon libraries were sequenced using the MiSeq Illumina system, and individual sequences were analyzed employing FastQC-v.0.11.8 software. Low-quality bases and adapter sequences were removed using the optimized settings of Trimmomatic v.0.39 (LEADING 3, TRAILING 3, SLIDINGWINDOW4 30, MINLEN 200). Phix control and host DNAs were removed from the pre-assembled data. Genomes were assembled using Spades v3.13.0 running the default settings. The assembled contigs were filtered by length lower than 1000 and k-mer coverage lower than 5. Single reliable genome sequences were identified.
The phylogenetic analysis was based on the F gene sequences of CD1901 and CD1901-100 and 23 other CDV strains from the GenBank database. Phylogenic tree construction using the neighbor-joining method of MEGA v.0.20 software (http://www.megasoftware.net/) was performed as described previously (16). CD1901/CD1901-100 amino acid sequence alignments were examined using Clone Manager Basic v.9 software (Sci-Ed Software, Denver, CO, USA).
Go to : 
RESULTS
Construction of Vero cells expressing the dog nectin-4 gene
We constructed a new Vero cell line expressing the dog nectin-4 gene for passage of CD1901. The dog nectin-4 gene was inserted into a lentiviral expression vector containing the GFP gene and the recombinant lentivirus used to infect normal Vero cells. As the infected cells expressed both the dog nectin-4 and GFP genes, fluorescence was apparent under a fluorescence microscope (Fig. 1) in the absence of any treatment. The two portions of the dog nectin-4 gene were PCR-identified in DNA of the Vero/dNectin-4 cells, because two primer sets were used (Fig. 2).
Biological characterization of CD1901-100
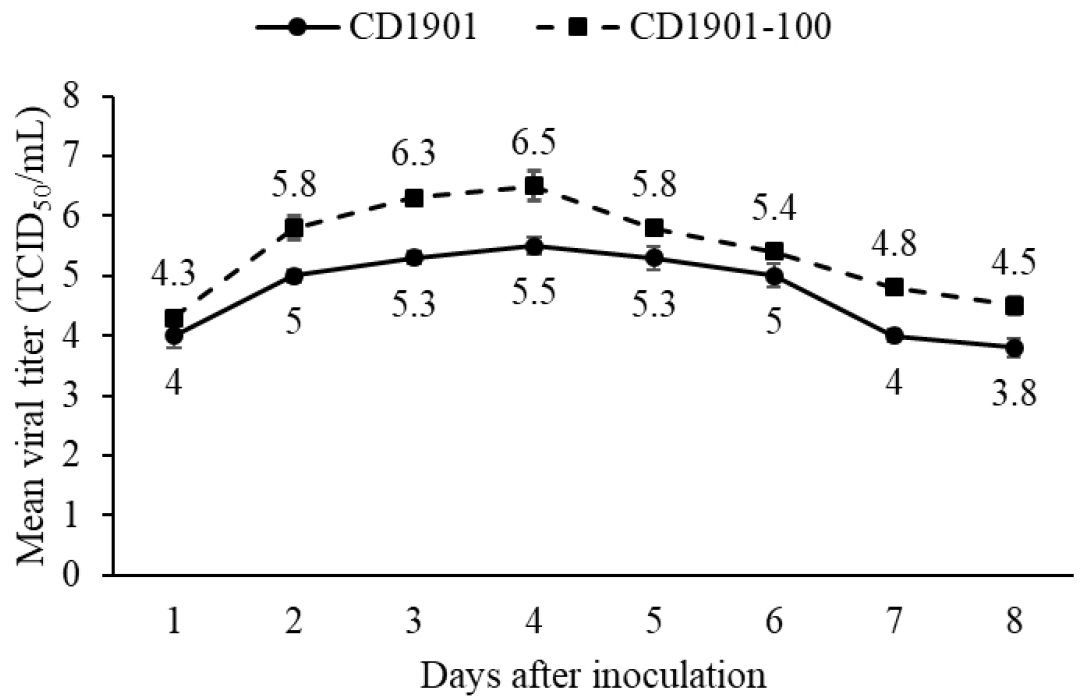
Vero/dSLAM cells infected with CD1901 presented as large round syncytia resembling giant cells (Fig. 3A). However, cells infected with CD1901-100 were broken, dispersed, and separated (Fig. 3B). Vero/dSLAM cells infected with CD1901 and CD1901-100 were fixed in cold acetone and stained with monoclonal antibodies against CDV. As shown in Figs. 3D and 3E, the fluorescence types differed, as did the CPEs of Vero/dSLAM cells infected with the CDVs. We also compared the viral growth kinetics to determine the optimal harvest time for CD1901-100 by reference to the CPEs induced by the CDVs. As shown in Fig. 4, for both CD1901 and CD1901-100, the highest viral titers of 105.5 and 106.5 TCID50/mL were attained at 4 day post inoculation. CD1901-100 was inoculated into seven types of cells. The highest viral titer of 107.0 TCID50/mL was afforded by Vero/dNectin-4 cells; in contrast, the titer was 106.6 TCID50/mL after growth in normal Vero cells. However, CD1901-100 grew to moderate or low titers ranging from 105.0 TCID50/mL to 102.6 TCID50/mL in DF-1, A72, and MDCK cells (Fig. 5).
 | Fig. 3Cytopathic effects (CPEs) and fluorescence images of Vero/dSLAM cells infected with CD1901 and CD1901-100. The CPEs and fluorescence varied among Vero/dSLAM cells infected with CD1901 (A, D) and CD1901-100 (B, E). No CPE or fluorescence was evident in normal Vero cells (C, F). Scale bars: 100 μm. |
Molecular characterization of CD1901-100
As shown in Table 3, the six open reading frames of CD1901-100 genome contained 14,472 nucleotides. Compared to the CD1901 genome, CD1901-100 had 25 nucleotide mutations and 15 amino acid substitutions. Of the six structural proteins, the F protein exhibited the highest amino acid replacement rate (5/663, 0.75%). As shown in Fig. 6, CD1901-100 had one amino acid substitution at position 484 of the H protein and had five replacements at positions 37, 39, 47, 124, and 279 of the F protein. Interestingly, four of the five substitutions were in the signal sequence of the F protein. A phylogenetic tree was constructed based on the F genes of the 25 CDV strains. As shown in Fig. 7, CD1901-100 belonged to the Asia-1 group and was most closely related to GS20-11 reported from China in 2021.
 | Fig. 6Alignments of partial amino acid sequences of the hemagglutinin (A) and fusion (B) proteins of CD1901 and CD1901-100. The potential signaling activation molecule-binding region of dog H protein (A) and the signal sequence of F protein (B) are shaded in gray. The one and five amino acid substitutions in the H and F proteins, respectively, are indicated in red. |
 | Fig. 7A phylogenetic analysis based on the nucleotide sequences of the complete Fusion (F) gene of 25 canine distemper virus strains. The CD1901 and CD1901-100 strains are of the Asia- 1 genotype and are closely related to the GS20-11 strain from China. The phylogenetic tree was constructed based on alignments of nucleotide sequences obtained using the neighbor-joining method. |
Table 3.
Mutations in six protein-encoding genes of CD1901-100
| Name of protein (Amino acid/nucleotide) | Nucleotide mutations (%)* | Amino acid substitutions (%)* |
|---|---|---|
| Nucleocapsid (N) (524/1,572) | 3 (99.8) | 3 (99.4) |
| Phosphoprotein (P) (508/1,524) | 5 (99.7) | 2 (99.6) |
| Matrix protein (M) (336/1,008) | 1 (99.9) | 1 (99.7) |
| Fusion protein (F) (663/1,989) | 9 (99.6) | 5 (99.2) |
| Hemagglutinin (H) (608/1,824) | 1 (99.9) | 1 (99.8) |
| Large protein (L) (2,185/6,555) | 6 (99.9) | 3 (99.9) |
| Total number of mutations (4,826/14,472) | 25 (14,447/14,472:99.8) | 15 (4,809/4,824:99.7) |
Go to : 
DISCUSSION
SLAM and nectin-4 expressed on the surfaces of lymphocytes and epithelial cells act as morbillivirus receptors (14, 16). Several cell lines expressing SLAM or nectin-4 have been used to attenuate wild CDVs via passage and to study CDV entry into cells (19, 20). Here, we constructed Vero cells expressing dog nectin-4 using a strategy similar to that employed to prepare Vero/dSLAM cells. Vero cells expressing dog nectin-4 were named Vero/dNectin-4 cells after confirming dog nectin-4 gene expression via PCR and GFP expression with fluorescence microscopy. As Vero/dSLAM cells have been used to isolate new CDVs of genotypes Asia-1 to-4, Vero/dNectin-4 cells along with other cells may be used to induce CDV adaptations that decrease neurovirulence (16).
Few reports on selection of inactivated CDV vaccine candidates via cell passage have appeared (21). Continuous CDV cell passage is time-consuming; new strategies for induction of mutations in pathogenic genes are required. Changes in viral biological features on cell passage are affected by the composition of medium and the nature of the cells. For example, three CDV strains that had been passaged 20 in Vero/dSLAM cells showed slight changes in growth and CPEs (21). We passaged CD1901 100 times, including 20 exposures to UV for 1 min and 10 exposures to 4 mM 5ʹ bromouracil. Four types of cells such as Vero/dSLAM, DF-1, Vero, and Vero/dNectin-4 cells were used over 100 passages because CD1901 proliferates in those types of cells. CD1901 and CD1901-100 differed significantly in terms of their CPEs, IFA results, growth kinetics, and cell tropism. Cells infected with CD1901-100 do not form syncytia and CD1901-100 can grow in A72, MDCK, Vero, and DF-1 cells. It is thus possible to differentiate CD1901-100 from field-type CDVs or vaccine viruses such as the Lederle, Onderstepport, and Rockborn strains (22). Reduced syncytial formation may reflect viral attenuation (23). The proliferative capacity of CD1901-100 increased from 105.5 TCID50/mL to 106.6 TCID50/mL in Vero/dSLAM cells, suggesting that CD1901-100 is a good candidate for a new inactivated CDV vaccine.
The lack of a proofreading function in RNA-dependent RNA polymerase allows RNA viruses to readily mutate (24). Morbilliviruses lacking effective proofreading by the L protein have developed many mutations during serial passages (24). Continuous CDV passage in cells treated with mutagens has mutated the codons of 26 amino acids (25). We subjected CD1901 to 100 serial passages, with exposure to 5ʹ bromouracil and UV light, in four types of cells. Base analogs, intercalating agents, and UV irradiation are known mutagens. UV irradiation and 5ʹ bromouracil, an analog of uracil, have been used previously to generate viral genetic variants (26, 27). The full nucleotide sequences of CD1901 and CD1901-100 were identified and the mutations compared. CD1901-100 exhibited 25 mutations and 15 amino acid substitutions in six structural proteins compared to CD1901, indicating that 5ʹ bromouracil, UV exposure, and serial passages in Vero/dSLAM, DF-1, Vero, and Vero/dNectin-4 cells mutagenized CD1901.
Several studies have found that amino acid substitutions within the H and F proteins affect fusion efficiency (23, 28). Alignment of the amino acid sequences of the H and F proteins revealed one and five amino acid substitutions in the CD1901 H and F proteins, respectively. Furthermore, there were one and four amino acid differences, respectively, in the SLAM binding sites of the H protein and the signal sequences of the F protein between the two CDVs, suggesting that such substitutions change syncytial formation in Vero/dSLAM cells.
Phylogenetic analyses of CDV genes have identified evolutionary relationships and determined the CDV genotypes (13). Sequencing of the F genes of 25 CDV strains revealed that CD1901 and CD1901-100 belong to the Asia-1 group and exhibit high-level nucleotide homology (99.5%), but both show lower homology (90.7%) to the Lederle strain. This large difference may explain the incomplete protection of dogs immunized with the current CDV vaccine. Chinese mink vaccinated with current commercial CDV vaccines based on the Synder Hill strain or the Onderstepoort strain were not protected against the WH2 CDV strain of the Asia-1 genotype (29). Thus, characterization of CD1901-100 with high viral titer was the first step in preparation of an inactivated CDV vaccine. Research on the immunogenicity and efficacy of the vaccine in target animals will be the second step.
In conclusion, several cells including Vero/dNectin-4 cells successfully propagated CD1901-100. This strain will serve as a new CDV vaccine strain for development of an inactivated CDV vaccine. Further studies on the immunogenicity of the new inactivated CDV vaccine in target animals are required.
Go to : 




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download