Abstract
Background
Carbapenemase-producing Enterobacterales (CPE) have been highlighted as an urgent threat by international health authorities. Rapid detection of carbapenemase will allow implementation of infection control measures.
Methods
We assessed the performance of the Xpert Carba-R assay (Cepheid, USA) designed for rapid detection of five most common carbapenemases (blaKPC, blaNDM, blaOXA-48, blaVIM, and blaIMP-1) from rectal swab specimens by comparing with the reference method, culture plus in-house PCR. A total of 26,178 specimens were collected from August 2018 to July 2020 and tested using the Xpert Carba-R assay and culture plus in-house PCR.
Results
Among the 26,178 specimens collected from 12,889 patients, 1,615 (6.2%) were detected using the Xpert Carba-R assay, whereas 1,525 specimens (5.8%) were detected using a reference method. The sensitivity, specificity, and positive and negative predictive values of the Xpert Carba-R assay compared to the results of the culture method were 97.4% (95% confidence interval [CI], 96.5% to 98.1%), 99.4% (95% CI, 99.3% to 99.5%), 91.8% (95% CI, 90.5% to 93.0%), and 99.8% (95% CI, 99.7% to 99.8%), respectively. The prevalence of CPE and carbapenem-resistant Enterobacteriaceae were 1.7% (220/12,889) and 5.4% (695/12,889), respectively. Klebsiella pneumoniae (108/220, 49.1%) was the most common species, followed by Escherichia coli (68/220, 30.9%) and Citrobacter freundii (17/220, 7.7%). blaKPC was the most common carbapenemase gene (123/220, 55.9%), followed by blaNDM (56/220, 25.4%), blaOXA-48 (27/220, 12.2%), and blaNDM/OXA-48 (9/220, 4.1%).
초록
배경
Carbapenemase-producing Enterobacterales (CPE)의 확산은 전 세계적으로 큰 위협이 되고 있으며 이들의 신속한 검출은 감염 확산 방지에 필수적이다.
방법
고위험환자군에서의 능동 감시를 위해 시행한 Xpert Carba-R assay (Cepheid, USA)의 5가지의 대표적인carbapenemase 유전자(blaKPC, blaNDM, blaOXA-48, blaVIM, blaIMP-1)의 검출 성능을 후향적으로 평가하였다. 2018년 8월부터 2020년 7월까지 내원한 총 12,889명 환자의 직장 도말 검체 26,178개를 참고 방법(증균배양과 in-house PCR로 유전자 확인)과 비교하였다.
결과
12,889명의 환자로부터 수집된 26,178개의 검체 중에서 1,615 검체(6.2%)가 Xpert Carba-R assay로 양성으로 확인되었으며, 1,525 검체(5.8%)가 참고 방법으로 양성으로 확인되었다. 참고 방법과 비교하여 민감도, 특이도, 양성 예측도 및 음성 예측도는 각각 97.4% (95% confidence interval [CI], 96.5%-98.1%), 99.4% (95% CI, 99.3%-99.5%), 91.8% (95% CI, 90.5%-93.0%) 및 99.8% (95% CI, 99.7%-99.8%)이었다. CPE와 CRE의 유병률은 각각 1.7% (220/12,889)와 5.4% (695/12,889)였다. 균 별로 Klebsiella pneumoniae (108/220, 49.1%)가 가장 많고, Escherichia coli (68/220, 30.9%)와 Citrobacter freundii (17/220, 7.7%) 순이었다. Carbapenemase 유전자 별로는 blaKPC가 가장 많았고(123/220, 55.9%), blaNDM (56/220, 25.4%), blaOXA-48 (27/220, 12.2%) 그리고 blaNDM/OXA-48 (9/220, 4.1%) 순이었다.
The rapid spread of carbapenemase-producing Enterobacterales (CPE) worldwide is a serious threat to public health [1], as it is capable of spreading resistance genes through horizontal transfer and has been related to several outbreaks [2]. In Korea, reporting CPE infections to the Korean Center for Disease control and Prevention (KCDC) has been made mandatory for hospitals since 2012 (http://www.kdca.go.kr/). According to the KCDC, there were 11,954 carbapenem-resistant Enterobacteriaceae (CRE) cases in 2018 and 15,369 cases in 2019. Among the 2019 cases, 57.8% (8,887/15,369) were confirmed as CPE infections [3]. For infection control, rapid identification of patients colonized with CPE has become a routine clinical aim in many parts of the world and is recommended by public health organizations [4]. Various methods to detect CPE in rectal swabs, including culture with specific chromogenic media and commercial molecular tests, have been used [5]. Although concomitant culture is necessary for bacterial species identification and epidemiological typing, it requires more than two days of turnaround time. To overcome this limitation, molecular tests have been developed with reduced processing time to rapidly implement proper infection control measures [6, 7]. As a rapid molecular test, the Xpert Carba-R assay is capable of detecting five carbapenemase genes (blaKPC, blaNDM, blaOXA-48, blaVIM, and blaIMP-1) in approximately one hour and has been used in our laboratory since July 2017 for active surveillance of CPE. In this study, we evaluated the performance of the Xpert Carba-R assay compared with the conventional method culture plus in-house PCR.
We retrospectively analyzed the routine test results of the Xpert Carba-R assay (Cepheid, Sunnyvale, CA, USA) and the conventional method culture plus in-house PCR from August 2018 to July 2020. Screening for CPE was performed using rectal swab specimens from patients with high risk of intestinal colonization with CPE in accordance with the infection control policy of Seoul St. Mary’s Hospital. Screening was performed for all patients admitted to the intensive care unit or the general ward if the patient had been transferred from another hospital after a stay longer than 1 week within the last 4 weeks. For all patients admitted to the intensive care unit, testing was performed at admission, after every subsequent seven days, and before discharge.
The rectal swab specimens were collected in eSwab 480CE (Copan Diagnostics, Inc., Brescia, Italy) and submitted to a clinical microbiology laboratory. The Xpert Carba-R assay and culture plus in-house PCR were performed simultaneously, as described below.
This study was approved by the Institutional Review Board of Seoul St. Mary’s Hospital, Seoul, South Korea (KC21RASI0674).
The rectal swabs were processed using Xpert Carba-R assay, a real-time PCR assay designed to detect the five most common carbapenemases, i.e., blaKPC, blaNDM, blaOXA-48, blaVIM, and blaIMP-1. The assay was performed according to the manufacturer's instructions. The transport media was sent to the laboratory and vortexed, and a 100 μL sample was mixed with Xpert Carba-R assay sample buffer. After vortexing the mixture, 1.7 mL of the sample buffer was transferred to an Xpert Carba-R assay cartridge and was placed on the GeneXpert™ system.
For conventional culture method, 100 μL of each specimen was placed in 8 mL of MacConkey broth (BD, Franklin Lakes, NJ, USA) containing a 10 μg ertapenem disk (BD). After 24 hours and 48 hours of incubation at 37°C, if the solution turned yellow or turbid, an aliquot of 100 μL was removed and inoculated onto MacConkey agar (Asan Pharmaceutical, Seoul, Korea) and CHROMagar mSuperCARBA agar (CHROMagar, Paris, France) plates. Next, 10 μg ertapenem disks were placed in the first and second quadrants of MacConkey agar for detection of carbapenem-resistant isolates. The colonies that grew within 27 mm of the ertapenem disks after overnight incubation at 37°C were suspected as CRE [8].
Additionally, for cases showing positive results in the Xpert Carba-R assay, 100 μL of each specimen was inoculated directly onto a CHROMagar plate for more rapid identification of CPE species. The species identification was performed using matrix assisted laser desorption ionization time-of-flight (MicroIDSys, ASTA, Suwon, Korea), and the antimicrobial susceptibility test was performed using Vitek-2 (bioMérieux, Marcy L’Etoile, France). To confirm the presence of carbapenemase genes, colonies were suspended in 1 mL distilled water in a 1.5 mL micro tube and vortexed. After washing, the pellet was boiled for 20 minutes for DNA extraction, followed by PCR using a C1000 Thermal Cycler Chassis (Bio-Rad, Hercules, CA, USA) to detect the most common five genes, blaSIM, and blaGES. The primer sequences used for PCR are described in previous studies [9, 10].
Growth of CPE either on MacConkey agar or CHROMagar with carbapenemase gene confirmation using in-house PCR was defined as true-positive. No growth of CPE was defined as true-negative. The sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) including 95% confidential interval (CI) were calculated for all test results [11]. Statistical analysis was performed using the MedCalc Statistical Software version 18 (MedCalc Software Bvba, Ostend, Belgium; http://www.medcalc.org; 2018). Differences in the threshold cycle (Ct) values between CPE and non-CPE (false-positive results of Xpert Carba-R assay) specimens were analyzed using the Mann-Whitney U-test. P values <0.05 were considered significant. Samples that found positive result with the Xpert Carba-R assay, 24 hours earlier-inoculation was done on CHROMagar directly. And the samples were compared with that inoculated on MacConkey broth. For analysis of prevalence, only one specimen per patient was used. The Chi-square test was used to compare culture results of specimens found to be true- or false-positives in the Xpert Carba-R assay. This test was also used to compare the results of the prevalence of CPE during the two periods.
A total of 26,178 specimens from 12,889 patients were tested from August 2018 to July 2020.
In the Xpert Carba-R assay, 6.2% positive cases (1,615/26,178 specimens) were detected. Furthermore, in the reference method, 5.8% positive cases (1,525/26,178 specimens) were detected (Table 1).
Table 2 shows the overall performance of the Xpert Carba-R assay compared to the reference method. Sensitivity, specificity, PPV, and NPV of the Xpert Carba-R assay were 97.4% (95% CI, 96.5% to 98.1%), 99.4% (95% CI, 99.3% to 99.5%), 91.8% (95% CI, 90.5% to 93.0%), and 99.8% (95% CI, 99.7% to 99.8%), respectively. There were 26,178 specimens analyzed for five individual genes, providing 130,890 test results. The results were defined as positive for the Xpert Carba-R assay if at least one carbapenemase gene was detected.
Table 3 shows discrepant results between the Xpert Carba-R assay and the reference method. Here, 39 cases that showed negative in the Xpert Carba-R assay but showed positive in the reference method were considered false-negative results of the Xpert Carba-R assay. Most of these were blaKPC-producing Klebsiella pneumoniae (N=29). In contrast, 131 specimens yielded false-positive results in the Xpert Carba-R assay. Among these, blaIMP-1 was the most common gene (N=48), followed by blaNDM, blaKPC, and blaOXA-48. In 90 of the 131 cases, CRE did not grow. In the remaining 41 cases, the carbapenemase genes were detected in non-fermentative bacilli, most of which were identified as Pseudomonas aeruginosa (23/41, 56.1%), followed by Acinetobacter baumannii (10/41, 24.4%).
Compared to the true-positive cases detected in the Xpert Carba-R assay, the median Ct value of false-positive cases was significantly higher (24.3 vs. 35.0, P<0.001, Mann-Whitney U-test) (Fig. 1). According to carbapenemase type, the median Ct values of true-positive groups were 25.4, 24.9, 23.3, 26.0, and 32.3 for blaKPC, blaNDM, blaOXA-48, blaVIM, and blaIMP-1, respectively. The median Ct values of false-positive groups were 29.0, 32.0, 34.1, 34.2, and 35.0 for blaKPC, blaNDM, blaOXA-48, blaVIM, and blaIMP-1, respectively. Differences in Ct values were statistically significant for all five genes, blaKPC (P=0.003), blaNDM (P<0.001), blaOXA-48 (P<0.001), blaVIM, (P=0.002), and blaIMP-1 (P=0.033).
Additionally, in the MacConkey broth culture, one blaGES-producing K. pneumoniae and two blaSIM-producing C. freundii were found using the in-house PCR method.
The prevalence of CPE and CRE were 1.7% (220/12,889) and 5.4% (695/12,889), respectively. Klebsiella pneumoniae (108/220, 49.1%) was the most common species, followed by E. coli (68/220, 30.9%) and C. freundii (17/220, 7.7%). blaKPC was the most common carbapenemase gene (123/220, 55.9%), followed by blaNDM (56/220, 25.4%), blaOXA-48 (27/220, 12.2%), and blaNDM/OXA-48 (9/220, 4.1%).
The prevalence of CPE from August 2018 to July 2020 is shown on Fig. 2. During the first period (2018.08-2019.07) of the study, the prevalence of CPE was 1.3% (87/6,546), and the proportion of CPE isolates among CRE isolates was 38.1% (87/228). Of these 87 isolates, 41 harbored the blaKPC gene, while the remaining isolates carried blaNDM (N=22) or blaOXA-48 (N=14) genes. Escherichia coli (37/87, 42.5%) was the most common species, followed by K. pneumoniae (25/87, 28.7%) and C. freundii (8/87, 9.2%). During the second period (2019.08-2020.07) of the study, the prevalence of CPE increased to 1.7% (133/6,343), and the proportion of CPE patients among CRE patients was 28.4% (133/467) due to the increase in the number of CRE patients. Of these 133 CPE cases, 82 harbored blaKPC, followed by the remaining cases carrying blaNDM (N=34) or blaOXA-48 (N=13). Klebsiella pneumoniae (83/133, 62.4%) isolates were the most common species during this period, followed by E. coli (31/133, 23.3%) and C. freundii (9/133, 6.8%). The most common species over the two years of the study showed different and increased blaKPC-producing K. pneumoniae up to 53.3% (71/133) from 20.6% (18/87). The results of the Chi-square test for carbapenemase gene and species showed P-value<0.05 for blaKPC-producing K. pneumonia; no other combination showed significant result.
The Xpert Carba-R assay detected 1,615 positive targets (827 blaKPC, 461 blaNDM, 371 blaOXA-48, 20 blaVIM, and 68 blaIMP-1) and multiple genes in 126 specimens (Table 1). Among multiple genes detected, blaNDM+blaOXA-48 were most common (73.8%, 93/126), consistent with previous studies in Korea [3, 12], and were followed by blaKPC+blaIMP-1 (6.3%, 8/126) and blaKPC+blaVIM (4.8%, 6/126). Species of blaNDM,+blaOXA-48 were E. coli, except for three specimens (two specimens for E. coli and Klebsiella oxytoca, and one specimen for E. coli and Kluyvera cryocrescens). In other multiple gene detected carbapenemases (33/126), blaKPC was the most common gene (25/33).
The Xpert Carba-R assay demonstrated high sensitivity and specificity (97.4% and 99.4%, respectively). Our results are in agreement with results from several studies showing excellent performance for each carbapenemase [13, 14]. The PPV for blaKPC was 97.4% in our study; however, Tato et al. [13] reported relatively low PPV for blaKPC (87.9%). Higher PPV in our study for blaKPC might be due to higher sensitivity of our reference method [15-17] or higher prevalence of blaKPC in the studied population. Higher sensitivity of CHROMagar than MacConkey agar with a meropenem disk for CPE screening was reported previously [5], and inclusion of CHROMagar as reference method might have increased the specificity and PPV of the Xpert Carba-R assay. This finding indicates that the high sensitivity of the reference method is critical for evaluation of molecular methods and that studies of the performance of the Xpert Carba-R assay can show differences in results [13, 18]. Additionally, 89 out of 1,484 CPE specimens showed negative results with the MacConkey broth after 24 hours incubation, while seven showed negative results with MacConkey broth after 48 hours incubation. Seven specimens were only confirmed using CHROMagar, four blaKPC-producing E. coli, one blaOXA-48-producing E. coli, and two blaKPC-producing K. pneumoniae. In contrast, no isolate was identified using MacConkey broth only. Therefore, inoculation of rectal swabs onto a CHROMagar plate allows faster and more accurate identification of CPE, which could prevent dissemination.
The PPV for blaIMP was 29.4%, which was lower than the PPV values of other carbapenemase genes, partly because of the low prevalence of blaIMP in Korea [3]. Additionally, blaIMP was mainly detected in non-fermentative bacilli, consistent with a previous study [19].
Among the 39 false-negative specimens, 29 harbored blaKPC-producing K. pneumoniae (Table 3). The small number of organisms in the rectal swab specimens might be the cause of false-negatives in these specimens. This is supported by the findings that most (94.0%, 1,395/1,484) of the true-positive specimens showed growth after 24 hours incubation in MacConkey broth, whereas as many as 30.8% (12/39) of the false-negative specimens showed growth after 48 hours incubation (Chi-square test, P-value<0.001).
Among the 131 false-positive results, growth of carbapenem-resistant isolates was not observed in 90 cases (68.7%). Among the remaining 41 cases, growth of P. aeruginosa and A. baumannii was observed in 23 and 10 cases, respectively (Table 3). A high median Ct value of 35.0 was found for the false-positive results (Fig. 1). This could be explained by a low level of target gene expression of the organism or the possibility of medicinal treatment or modified sequence of the target gene. Compared to the true-positive cases detected using the Xpert Carba-R assay, the median Ct value of false-positive cases was significantly higher (24.3 vs. 35.0, P<0.001) (Fig. 1).
In our study, rare carbapenemase genes (blaSIM and blaGES) were detected from three isolates; the blaSIM was found in two C. freundii isolates, while the blaGES was found in one K. pneumoniae isolate. The prevalence of blaGES and blaSIM genes was very low in our study (0.2%), in line with the population results of studies conducted in Korea, the United States, and Europe [12, 20]. However, it is essential to verify the absence of blaGES and blaSIM genes to prevent dissemination through horizontal spread. A recent study in India about dissemination of plasmids encoding carbapenemases in gram-negative bacteria showed blaGES as the third most common gene (11 of 37, 29.7%, 6 for blaGES-1 and 5 for blaGES-9) in E. coli.
One of the limitations of our study is that we did not perform a direct PCR or sequencing of the MacConkey broth. Tato et al. [13] demonstrated that 11 of 18 specimens previously classified as false-positives using the Xpert Carba-R assay were reclassified as true-positives after discrepant sequencing analysis of the MacConkey broth. This might be due to low bacterial load of carbapenemase in the rectal swab specimens or failure of carbapenemase-harboring species to recover using the initial culture method. And Our study did not include the new emerging blaOXA variants, such as blaOXA-232 [21].
However, to the best of our knowledge, this study included the largest number of clinical specimens. Based on the results obtained, we confirmed high NPV (99.8%) of the Xpert Carba-R assay, with the prevalence of up to 27%. Considering the high sensitivity (97.4%) and NPV (99.8%) of the Xpert Carba-R assay, for cases showing culture positivity, confirmatory PCR might be omitted in regions with a very low prevalence of other carbapenemases not included in the Xpert Carba-R assay such as blaGES, and only species identification and antimicrobial susceptibility test would be needed for identification of CRE.
In conclusion, the prevalence of both CPE and CRE was higher in 2020 compared to that in 2019. The Xpert Carba-R assay showed excellent performance and can be used as a useful tool for active surveillance for CPE.
REFERENCES
1. Albiger B, Glasner C, Albiger B, Glasner C, Struelens MJ, Grundmann H, et al. 2015; the European Survey of Carbapenemase-Producing Enterobacteriaceae (EuSCAPE) working group. Carbapenemase-producing Enterobacteriaceae in Europe: assessment by national experts from 38 countries, May 2015. Euro Surveill. 20:30062. DOI: 10.2807/1560-7917.ES.2015.20.45.30062. PMID: 26675038.
2. Tokatlidou D, Tsivitanidou M, Pournaras S, Ikonomidis A, Tsakris A, Sofianou D. 2008; Outbreak caused by a multidrug-resistant Klebsiella pneumoniae clone carrying blaVIM-12 in a university hospital. J Clin Microbiol. 46:1005–8. DOI: 10.1128/JCM.01573-07. PMID: 18199780. PMCID: PMC2268336.

3. Korea Centers of Disease Control, Prevention. Incidence of carbapenem-resistant enterobacteriae in Korea, 2018-2020. https://www.kdca.go.kr/board/board.es?mid=a20602010000&bid=0034&act=view&list_no=717046.
4. Centers for Disease Control, Prevention. 2015. Facility guidance for control of carbapenem-resistant Enterobacteriaceae (CRE): November 2015 update - CRE toolkit 2015. https://www.cdc.gov/hai/pdfs/cre/CRE-guidance-508.pdf. Updated on Nov 2015.
5. Viau R, Frank KM, Jacobs MR, Wilson B, Kaye K, Donskey CJ, et al. 2016; Intestinal carriage of carbapenemase-producing organisms: current status of surveillance methods. Clinical Microbiol Rev. 29:1–27. DOI: 10.1128/CMR.00108-14. PMID: 26511484. PMCID: PMC4771221.

6. Nijhuis R, Samuelsen Ø, Savelkoul P, van Zwet A. 2013; Evaluation of a new real-time PCR assay (Check-Direct CPE) for rapid detection of KPC, OXA-48, VIM, and NDM carbapenemases using spiked rectal swabs. Diagn Microbiol Infect Dis. 77:316–20. DOI: 10.1016/j.diagmicrobio.2013.09.007. PMID: 24135412.

7. Braun SD, Monecke S, Thürmer A, Ruppelt A, Makarewicz O, Pletz M, et al. 2014; Rapid identification of carbapenemase genes in gram-negative bacteria with an oligonucleotide microarray-based assay. PLoS One. 9:e107079. DOI: 10.1371/journal.pone.0107079. PMCID: PMC4152334.

8. Lolans K, Calvert K, Won S, Clark J, Hayden MK. 2010; Direct ertapenem disk screening method for identification of KPC-producing Klebsiella pneumoniae and Escherichia coli in surveillance swab specimens. J Clin Microbiol. 48:836–41. DOI: 10.1128/JCM.01988-09. PMID: 20071553. PMCID: PMC2832415.

9. Poirel L, Walsh TR, Cuvillier V, Nordmann P1. 2011; Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 70:119–23. DOI: 10.1016/j.diagmicrobio.2010.12.002. PMID: 21398074.

10. Bradford PA, Bratu S, Urban C, Visalli M, Mariano N, Landman D, et al. 2004; Emergence of carbapenem-resistant Klebsiella species possessing the class A carbapenem-hydrolyzing KPC-2 and inhibitor-resistant TEM-30 beta-lactamases in New York City. Clin Infect Dis. 39:55–60. DOI: 10.1086/421495. PMID: 15206053.

11. Clark RB, Sharp SB. 2009. Cumitech 31A: Verification and validation of procedures in the clinical microbiology laboratory. Washington, DC: American Society for Microbiology.
12. Park SH, Kim JS, Kim HS, Yu JK, Han SH, Kang MJ, et al. 2020; Prevalence of carbapenem-resistant Enterobacteriaceae in Seoul, Korea. J Bacteriol Virol. 50:107–16. DOI: 10.4167/jbv.2020.50.2.107.
13. Tato M, Ruiz-Garbajosa P, Traczewski M, Dodgson A, McEwan A, Humphries R, et al. 2016; Multisite evaluation of Cepheid Xpert Carba-R assay for detection of carbapenemase-producing organisms in rectal swabs. J Clin Microbiol. 54:1814–9. DOI: 10.1128/JCM.00341-16. PMID: 27122379. PMCID: PMC4922077.

14. Moore NM, Cantón R, Carretto E, Peterson LR, Sautter RL, Traczewski MM. Carba-R Study Team. 2017; Rapid identification of five classes of carbapenem resistance genes directly from rectal swabs by use of the Xpert Carba-R assay. J Clin Microbiol. 55:2268–75. DOI: 10.1128/JCM.00137-17. PMID: 28515213. PMCID: PMC5483930.

15. Logan LK, Weinstein RA. 2017; The epidemiology of carbapenem-resistant Enterobacteriaceae: the impact and evolution of a global menace. J Infect Dis. 215:S28–36. DOI: 10.1093/infdis/jiw282. PMID: 28375512. PMCID: PMC5853342.

16. Tenny S, Hoffman MR. Prevalence. https://www.ncbi.nlm.nih.gov/books/NBK430867/. Updated on May 2021.
17. Pham HP, Staley EM, Raju D, Marin MJ, Kim CH. 2020; Laboratory assay evaluation demystified: a review of key factors in˝uencing interpretation of test results using different assays for SARS-CoV-2 infection diagnosis. Lab Med. 51:e66–70. DOI: 10.1093/labmed/lmaa045. PMID: 32634229. PMCID: PMC7454829.
18. Byun J-H, Kim YA, Kim M, Kim B, Choi JY, Park YS, et al. 2020; Evaluation of Xpert Carba-R assay v.2 to detect carbapenemase genes in two hospitals in Korea. Ann Lab Med. 40:209–15. DOI: 10.3343/alm.2020.40.3.209. PMID: 31858760. PMCID: PMC6933065.

19. Hong DJ, Bae IK, Jang IH, Jeong SH, Kang HK, Lee K. 2015; Epidemiology and characteristics of metallo-β-lactamase-producing Pseudomonas aeruginosa. Infect Chemother. 47:81–97. DOI: 10.3947/ic.2015.47.2.81. PMID: 26157586. PMCID: PMC4495280.

20. van Duin D, Doi Y. 2017; The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence. 8:460–9. DOI: 10.1080/21505594.2016.1222343. PMID: 27593176. PMCID: PMC5477705.

21. Jeong SH, Lee KM, Lee J, Bae IK, Kim JS, Kim HS, et al. 2015; Clonal and horizontal spread of the blaOXA-232 gene among Enterobacteriaceae in a Korean hospital. Diagn Microbiol Infect Dis. 82:70–2. DOI: 10.1016/j.diagmicrobio.2015.02.001. PMID: 25702524.
Fig. 1
Comparison of median Ct value and interquartile range in (A) total true-positive (24.4, 20.4-28.8) and false-positive (35.1, 32.3-36.8) cases, (B) each true/false-positive carbapenemase type, KPC (24.7, 21.2-29.3), KPC* (29.9, 23.6-33.6), NDM (24.4, 20.6-29.2), NDM* (33.2, 30.7-34.4), OXA-48 (22.7, 19.1-26.9), OXA-48* (35.0, 32.1-37.0), VIM (25.1, 21.9-32.1), VIM* (34.2, 34.1-36.1), IMP (33.2, 28.1-35.9), and IMP* (35.4, 33.9-36.8). Each boxplot represents the range from the 25th to the 75th percentiles of each group.
*carbapenemase genes with/without the star symbol indicate false/true-positive cases as observed in the Xpert Carba-R assay.
Abbreviations: TP, true-positive; FP, false-positive.

Fig. 2
Distribution of genotypes of CPE for two periods: (A) August, 2018-July, 2019 and (B) August, 2019-July, 2020.
Abbreviations: KPN, K. pneumoniae; ECO, E. coli; CFR, C. freundii; KOX, K. oxytoca; ECL, E. cloacae.
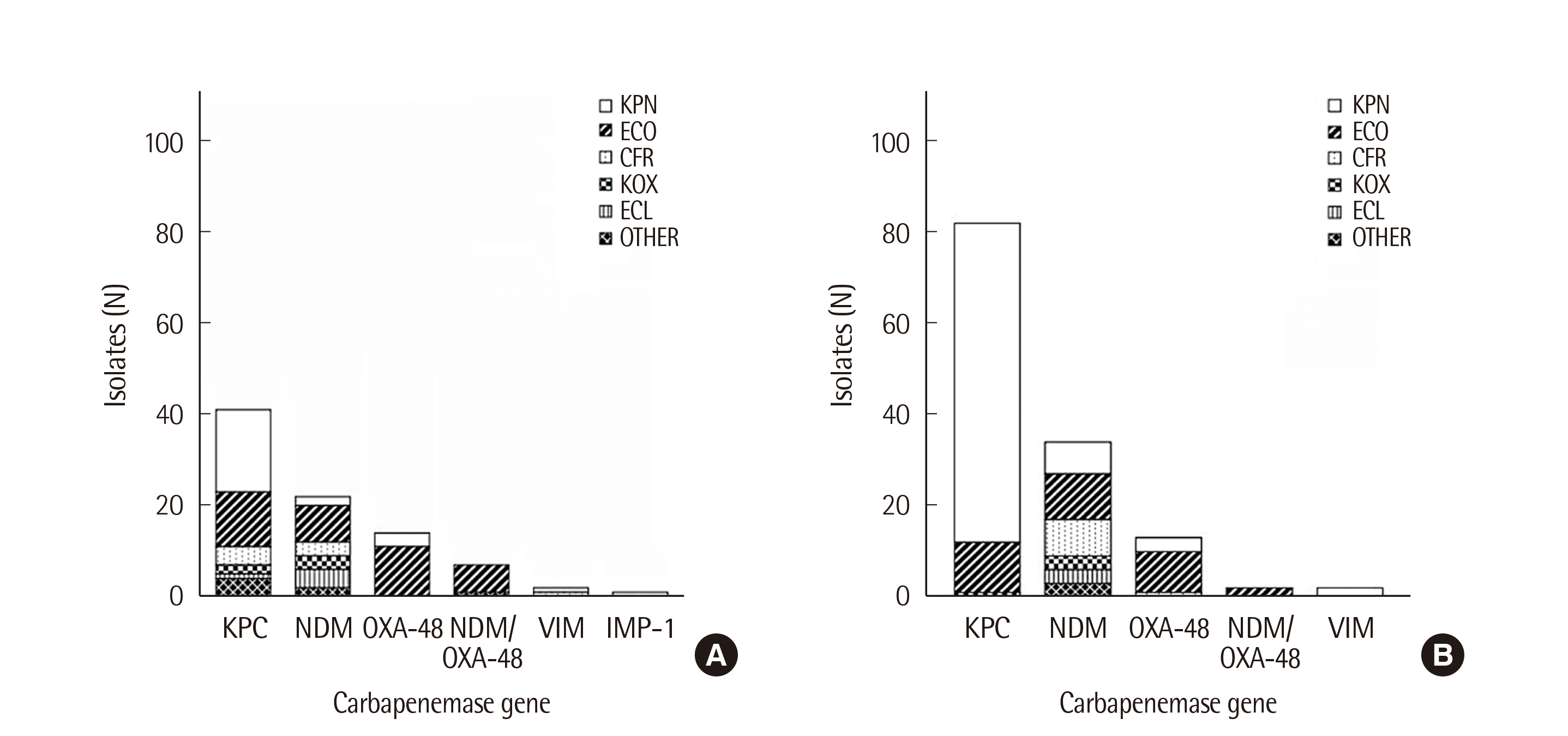
Table 1
Xpert Carba-R assay results by target gene for clinical specimens
| Carbapenemase gene | Clinical specimens (N=26,178) | ||
|---|---|---|---|
|
|
|||
| Xpert Carba-R assay | Reference method (Culture plus in-house PCR) | ||
| Single target gene | Positive | 1,615 | 1,525 |
| KPC | 802 | 811 | |
| NDM | 353 | 324 | |
| OXA-48 | 277 | 262 | |
| IMP | 52 | 4 | |
| VIM | 5 | 4 | |
| Total | 1,489 | 1,405 | |
| Combined target genes | NDM+OXA48 | 93 | 93 |
| KPC+IMP | 8 | 8 | |
| KPC+VIM | 6 | 6 | |
| KPC+NDM | 5 | 5 | |
| KPC+NDM+VIM | 4 | 4 | |
| NDM+IMP | 3 | 2 | |
| IMP+VIM | 3 | 0 | |
| KPC+NDM+IMP | 1 | 1* | |
| KPC+OXA48 | 1 | 1 | |
| NDM+VIM | 1 | 0 | |
| NDM+IMP+VIM | 1 | 0 | |
| Total | 126 | 120 | |
| Negative | 24,563 | 24,653 | |
Table 2
Overall Xpert Carba-R assay performance compared to the reference method (culture plus in-house PCR) for clinical specimens
Table 3
Discrepancy analysis of the Xpert Carba-R assay and the reference method (culture plus in-house PCR)
| Xpert Carba-R Assay | True results | Reference method (culture plus in-house PCR) | |||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|||||||||
| KPN | ECO | PAE | ABA | Other* | NG | Total | |||
| FN | KPC | 29 | 1 | 30 | |||||
| OXA-48 | 3 | 3 | 6 | ||||||
| NDM | 1 | 1 | 2 | ||||||
| VIM | 1 | 1 | |||||||
| Total | 34 | 5 | 39 | ||||||
| FP | IMP-1 | Negative | 9 | 5 | 4 | 30 | 48 | ||
| NDM | Negative | 11 | 5 | 18 | 34 | ||||
| OXA-48 | Negative | 2 | 15 | 21 | |||||
| KPC | Negative | 4 | 21 | 21 | |||||
| VIM | Negative | 1 | 6 | 7 | |||||
| Total | 23 | 10 | 8 | 90 | 131 | ||||




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download