Systemic mastocytosis (SM) is characterized by the clonal neoplastic proliferation of mast cells accumulating in one or more organ systems [
1]. SM with associated hematological neoplasm (SM-AHN) poses diagnostic challenges because of the coexistence of non-mast cell lineage hematological malignancies, including myelodysplastic syndrome (MDS), myeloproliferative neoplasm (MPN), and acute myeloid leukemia (AML), which mask the SM components [
1-
4]. We evaluated SM-AHN in patients with AML with
RUNX1::RUNX1T1 as they are known to have a high prevalence of
KIT variants, which are one of the diagnostic criteria for SM-AHN.
SM diagnosis depends on histopathological findings and the identification of mast cell aggregates, especially via immunohistochemical (IHC) staining, where mast cell aggregates are identified by CD117 staining and aberrant CD25 with or without CD2 expression indicate SM [
3,
5].
KIT variants detected in SM are often simultaneously found in AHN, especially in core-binding factor (CBF)-AML: 35% in AML with inv(16)(p13.1q22);
CBFB::MYH11 and 25% in AML with
RUNX1::RUNX1T1 have
KIT variants [
6,
7]. As the
KIT D816V variant is a minor criterion for SM-AHN diagnosis, patients diagnosed as having AML with inv(16) or AML with
RUNX1::RUNX1T1 may have concurrent SM-AHN [
2,
8,
9].
KIT variants are present in both mast cells and myeloid blasts [
10]. The variant burden has prognostic significance in advanced SM, including SM-AHN [
11].
Although cases of masked SM-AHN with AML have been previously reported, IHC staining of CD117 and CD25 for differential diagnosis are not routinely included in the work-up of SM-AHN in AML with RUNX1::RUNX1T1. We assessed the clinical significance of SM-AHN in patients with AML with RUNX1::RUNX1T1, focusing on IHC staining results and KIT variant detection after induction chemotherapy. As mast cells are enriched in tissues, we investigated whether the variant burden differs based on the sample type (bone marrow (BM) aspirate, a smear slide, or trephine biopsy tissue) and evaluated the changes in these differences post induction therapy.
The presence of SM-AHN was assessed in patients diagnosed as having AML with
RUNX1::RUNX1T1 in Seoul National University Bundang Hospital, Seongnam, Korea, from December 2014 to April 2020, and SM-AHN was diagnosed according to WHO criteria [
1]. This study was approved by the Institutional Review Board of Seoul National University Bundang Hospital (B-1711/435-004, B-2005/612-106) and Seoul National University Hospital (J-2108-080-1245). Informed consent was obtained from those patients in whom additional studies were performed. CD117 and/or CD25 IHC staining results at diagnosis, post induction and/or follow-up, were compared in patients with SM-AHN.
KIT variants were detected in BM aspirates by Sanger sequencing and/or next-generation sequencing (NGS). NGS was performed on BM aspirates of patients when requested by the clinician, using the Oncomine Myeloid Research Assay (Thermo Fisher, Waltham, MA, USA), IonS5 XL (Thermo Fisher), or a customized panel with the NextSeq 550 platform (Illumina, San Diego, CA, USA).
The KIT D816V variant burden in different samples, including BM aspirates, smear slides, and trephine biopsy sections, was evaluated at diagnosis and follow-up using droplet digital PCR (ddPCR). DNA was extracted using a QIAamp DNA Blood Mini kit (Qiagen, Hilden, Germany) and QIAcube (Qiagen). ddPCRs were run in a QX200 Droplet Digital PCR System (Bio-Rad, Hercules, CA, USA) according to the manufacturer’s instructions. The thermal cycles were as follows: enzyme activation at 95°C (10 minutes); 40 cycles of denaturation at 94°C (30 seconds), annealing and extension at 55°C (1 minute), and enzyme inactivation at 98°C (10 minutes); and holding at 4°C. Gating was performed based on positive and negative controls and analyzed using the QuantaSoft software v1.7.4 (Bio-Rad). Data were considered valid when at least 15,000 droplets were available.
Survival analysis was performed using the Kaplan-Meier method to compare overall survival (OS) in AML with RUNX1::RUNX1T1 with or without SM-AHN. OS was defined as the time from the date of diagnosis to the date of death or last follow-up. Categorical values were compared using Fisher’s exact test, and continuous variables were compared using the Mann-Whitney test. Statistical analyses were performed using SPSS version 25 (IBM Corporation, Armonk, NY, USA) and Prism 9.3.1 (GraphPad, San Diego, CA, USA).
Twenty-three patients were diagnosed as having AML with
RUNX1::RUNX1T1 during the study period, four of whom (17.4%) were also diagnosed as having SM-AHN. The clinical characteristics of the patients with or without SM-AHN are summarized in
Table 1. No significant differences in clinical characteristics were observed between the two groups, except for the presence of
KIT variants (
P=0.040).
KIT variants were present in six (26.1%) AML patients with
RUNX1::RUNX1T1, in three out of four patients with SM-AHN (75%; two D816V and one N822K) and in three patients without SM-AHN (15.8%; D816V, A820G, Y418delinsSVYIYIH). Two (50%) patients with D816V
KIT variants were diagnosed as having SM-AHN. Patients with or without SM-AHN did not show a significant difference in OS (
P=0.565).
Table 1
Characteristics of patients with AML with RUNX1::RUNX1T1
|
Characteristics |
Without SM-AHN (N = 19) |
With SM-AHN (N = 4) |
P
|
|
Age (yr) |
|
|
|
|
Median (range) |
43 (4–74) |
38.5 (8–56) |
0.683 |
|
Sex, N (%) |
|
|
|
|
Male |
12 (63.2) |
2 (50) |
1.000 |
|
Female |
7 (36.8) |
2 (50) |
|
|
Hb (g/L) |
|
|
|
|
Median (range) |
84 (52–116) |
86 (69–101) |
0.892 |
|
Leukocytes ( × 109/L) |
|
|
|
|
Median (range) |
5,810 (750–26,610) |
10,540 (4,690–29,930) |
0.366 |
|
Absolute neutrophil count ( × 109/L) |
|
|
|
|
Median (range) |
1,243 (66–13,017) |
730 (125–1,373) |
0.457 |
|
Platelets ( × 109/L) |
|
|
|
|
Median (range) |
57 (22–110) |
61 (16–81) |
0.953 |
|
KIT variant, N (%) |
3 (15.8) |
3 (75) |
0.040 |
|
D816V |
2 (10.5) |
2 (50) |
|
|
Other |
1 (5.3) |
1 (25) |
|
|
Outcome |
|
|
|
|
Follow-up, months, median (range) |
22 (2–68) |
33 (11–70) |
0.494 |
|
Hematopoietic stem cell transplantation, N (%) |
11 (57.9) |
4 (100) |
0.257 |
|
Death, N (%) |
2 (11.1) |
1 (25.0) |
0.470 |

We evaluated whether SM-AHN and AML diagnoses were concurrent. All patients were diagnosed as having SM-AHN based on follow-up BM tests. Patients with SM-AHN at the time of AML diagnosis did not show evident mast cell aggregation on hematoxylin-eosin slides. Thus, IHC staining for CD117 and CD25 was performed for diagnostic and follow-up BM samples (
Fig. 1). In retrospective analysis, CD117 expression was observed in mast cell aggregates at diagnosis in two patients, whereas it was not evident in the other two patients, with diffuse CD117-positive staining observed on the blasts at diagnosis of AML. Aberrant CD25 expression on mast cells at diagnosis was evident in only one patient. However, IHC staining of BM samples after induction revealed the presence of mast cell aggregates with CD117 expression and aberrant CD25 expression in all patients.
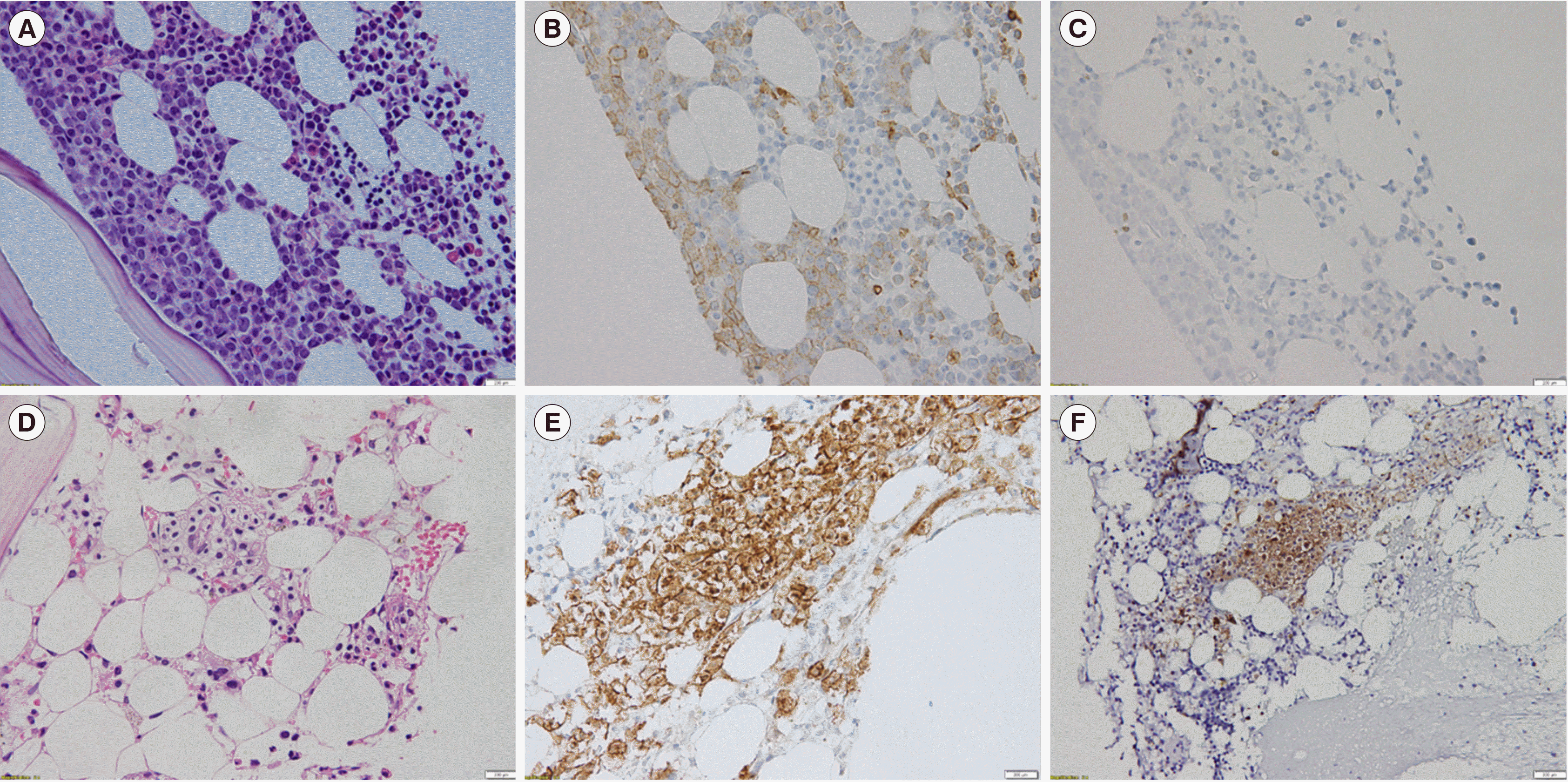 | Fig. 1
BM biopsy section and IHC staining results at diagnosis showing an increase in blasts and scattered mast cells (A, B, C) and evident mast cell aggregates post induction (D, E, F). (A, D) Hematoxylin-eosin stain (×400). (B, E) CD117 positivity in mast cells (×400). (C, F) Aberrant CD25 expression in atypical mast cells (×400).
Abbreviations: BM, bone marrow; IHC, immunohistochemical.

|
The
KIT D816V variant burden in two patients with SM-AHN who had
KIT D816V was assessed using diagnostic and follow-up samples (
Table 2). The variant allele burden decreased with decreasing blast count and was similar in all three sample types. The variant burden was low in post-induction samples and lower in BM aspirates than in smear slides and tissue sections for both patients. However, the
KIT D816V variant persisted after induction. Targeted sequencing results were obtained for two patients with SM-AHN and revealed additional variants in
RUNX1 (N=2), FLT3 (N=1), TP53 (N=1), and
NRAS (N=1) at diagnosis.
Table 2
ddPCR results for KIT D816V in BM aspirates, smear slides, and tissue sections
|
Sample number |
Sampling time |
Sample type |
Blast count (%) |
Mutant allele burden (%) |
|
S1 |
Diagnosis |
BM smear slide |
52.2 |
46.5 |
|
|
BM aspirate |
|
46.9 |
|
|
BM tissue |
|
47.3 |
|
Post-induction |
BM smear slide |
0.5 |
1.3 |
|
|
BM aspirate |
|
0.6 |
|
|
BM tissue |
|
1.2 |
|
S2 |
Diagnosis |
BM smear slide |
80.7 |
32.1 |
|
|
BM aspirate |
|
31.8 |
|
|
BM tissue |
|
50.0 |
|
Post-induction |
BM smear slide |
2.6 |
3.2 |
|
|
BM aspirate |
|
0.1 |
|
|
BM tissue |
|
3.2 |

Concomitant SM-AHN was diagnosed in 17.4% of the AML patients with
RUNX1::RUNX1T1 via CD117 and CD25 IHC staining in post-induction BM sections. Many studies have reported SM-AHN diagnosis in patients with AML [
4,
8,
12,
13]; however, the clinical characteristics of patients with and without SM-AHN have not been compared. The OS and clinical characteristics did not differ significantly, but
KIT variants were more prevalent in patients with SM-AHN. SM-AHN is difficult to diagnose in AML with
RUNX1::RUNX1T1 because of a marked expansion of blasts and CD117 expression in blasts [
4]. As NGS is routinely performed as a diagnostic test for myeloid malignancies, the presence of
KIT D816V has been reported to predict concurrent SM-AHN in MDS, MPN, and AML [
9]. A higher
KIT variant burden in tissue samples than in liquid samples has been reported in mastocytosis, including advanced SM [
11]. In our study, although only two patients were tested, the variant allele frequency in the BM smear slides was similar to that in the BM tissues, but greater than that in the BM aspirates, because BM components are enriched in tissues and smear slides. Thus, when fresh BM aspirate is not available, archived BM smear slides can serve as a reliable source of DNA for variant analysis [
14]. The genomic analysis of AML with
RUNX1::RUNX1T1 showed that the
KIT D816V variant was present in 17.4% of patients (N=4). As these patients tested positive for one minor criterion, it is advisable to screen for mast cell collections. However, according to the European Competence Network on Mastocytosis (ECNM), non-D816V
KIT variants are present in <10% of patients with mastocytosis, and therefore, the ECNM recommends whole
KIT gene sequencing when
KIT variant is not detected by codon 816-targeted assays, and non-D816V
KIT variants may show a different response to treatment [
15-
17]. The persistence of
KIT variants after induction therapy may be attributed to the SM or AHN component. Further, neoplastic mast cells of SM associated with AML with t(8;21) also carried
RUNX1::RUNX1T1 as well as
KIT variant as indicated by targeted FISH and single-cell analysis [
10,
18]. Thus, it is difficult to predict the SM burden or use
KIT variant burden as a minimal residual disease marker for SM-AHN [
9]. Since therapies targeting the SM component of SM-AHN are available, confirmation of the diagnosis is clinically significant, especially for advanced SM [
17,
19]. SM-AHN diagnosis requires additional testing with IHC staining after induction, and testing for
KIT variants may be performed using various BM samples with high mutation burdens, such as tissues or smear slides. Although a previous report has suggested that SM-AHN is uncommon in patients with CBF-AML, in this study, SM-AHN diagnosis was confirmed in 50% of the patients with
KIT-mutated AML with
RUNX1::RUNX1T1 [
20]. Therefore, it is necessary to screen mast cell aggregates for CD117 expression and subsequently for CD25 expression through IHC staining of samples collected from AML patients with
RUNX1::RUNX1T1 after induction.
Go to :






 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download