INTRODUCTION
Measurement uncertainty (MU) is a concept commonly used in various industries and engineering fields but not in clinical laboratories [
1]. As the importance of standardization and traceability of test results is increasing, MU is likely to become an important issue in laboratory quality management [
2-
4].
Since the Guide to the Expression of Uncertainty in Measurement (GUM) was published in 1996 [
5], the bottom-up approach is the standard method for estimating MU. This approach involves the identification of all sources of uncertainty in the measurement procedure, estimation of their magnitudes, and calculation of the combined uncertainty according to the law of error propagation [
5]. However, the MU guidelines for clinical laboratories recommend that the top-down (TD) approach is practical and particularly well-suited to closed measuring systems, which are common in routine clinical laboratories [
6,
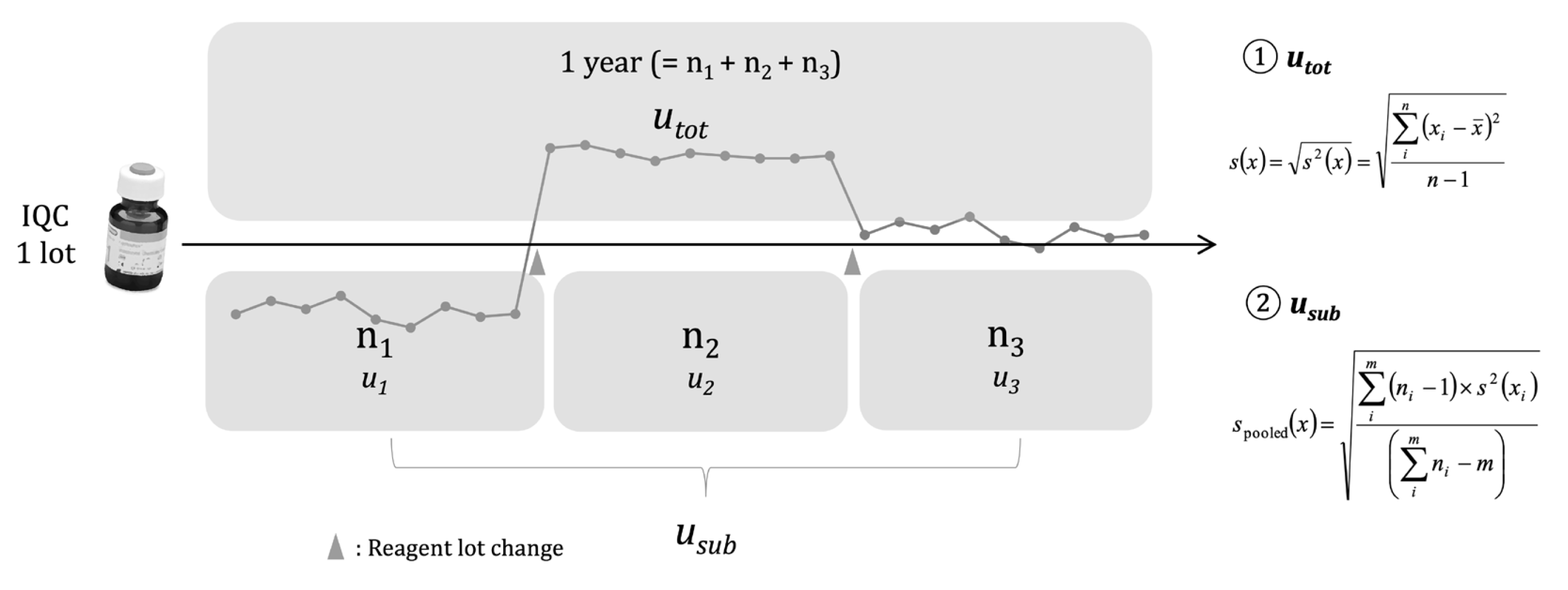
7]. For most measuring systems in clinical laboratories, the most significant uncertainty contributions to the overall MU are (1) long-term imprecision data obtained for internal quality control (IQC) materials for a period sufficient to include all changes to measuring conditions (
uRw, within-laboratory reproducibility); (2) uncertainty of the end-user calibrator (
ucal) obtained from the manufacturer or established by a laboratory with its own measuring system; and (3) bias correction, if a medically unacceptable measurement bias exists [
4,
6,
7].
Identifying the sources of uncertainty may be the first step in estimating the MU of a measurement system. Various measurement factors, such as sample inhomogeneity, reconstitution procedures for lyophilized materials, reagent and calibrator instability, fluctuations in the laboratory environment, operator bias, routine instrument maintenance, lot changes for calibrators and reagents, and different operators, are common sources of MU [
5-
9]. It is presumed that IQC data cover all anticipated routine changes in the measuring system for an appropriate period [
6,
7,
10]. When repeatability or long-term imprecision data for a well-controlled measurement procedure are plotted as a Gaussian distribution, the magnitude of the dispersion of values around the mean value can be quantified by calculating the standard deviation (SD) [
6,
7,
10]. Standard uncertainty can be expressed as SD. Because SD or
u values cannot be added or subtracted, relative standard uncertainties (
urel) first have to be converted to their respective variances (SD
2 and CV
2) in calculations [
6,
7,
10].
Among the various MU factors mentioned above, reagent lot changes are important factors that may cause a shift in IQC values, leading to MU overestimation [
7]. Therefore, it is recommended that both IQC and human sample results demonstrate similar behaviors upon a reagent lot change [
7]. If IQC values obtained before and after lot change are treated as a single dataset for
uRw calculation, MU may be overestimated. Practical considerations for when a shift occurs after a reagent lot change are reported in several guidelines [
7]. However, more specific recommendations are needed, e.g., a “significant change” upon a reagent lot change has to be clearly defined. In addition, the extent of differences that such considerations can bring about when MU is calculated using real-world IQC data should be demonstrated.
In this regard, we estimated MU by the TD approach using long-term IQC data generated in our laboratory to demonstrate how reagent lot changes influence uncertainty.
DISCUSSION
Guidelines issued by the International Organization for Standardization (ISO) and the CLSI recommend the TD approach for MU estimation in clinical laboratories [
6-
8]. In this approach, IQC data, which are easily obtainable in clinical laboratories, are required as a key component to evaluate MU, particularly, in laboratories that use closed measurement systems [
14]. However, the practical issues faced in working conditions need to be addressed. For example, the recommended collection period, described as a “sufficiently long time,” should be defined in detail [
6,
7]. Furthermore, clear recommendations should be made for IQC data obtained from different reagent lots. The guidelines recommend collecting IQC data separately if “a significant shift” in the IQC absolute values occurs when a new lot of reagents is introduced [
6,
7]. However, the range of acceptable values is not defined. If each laboratory uses different standards to calculate MU, the accuracy of the results may be compromised and/or confusion may arise.
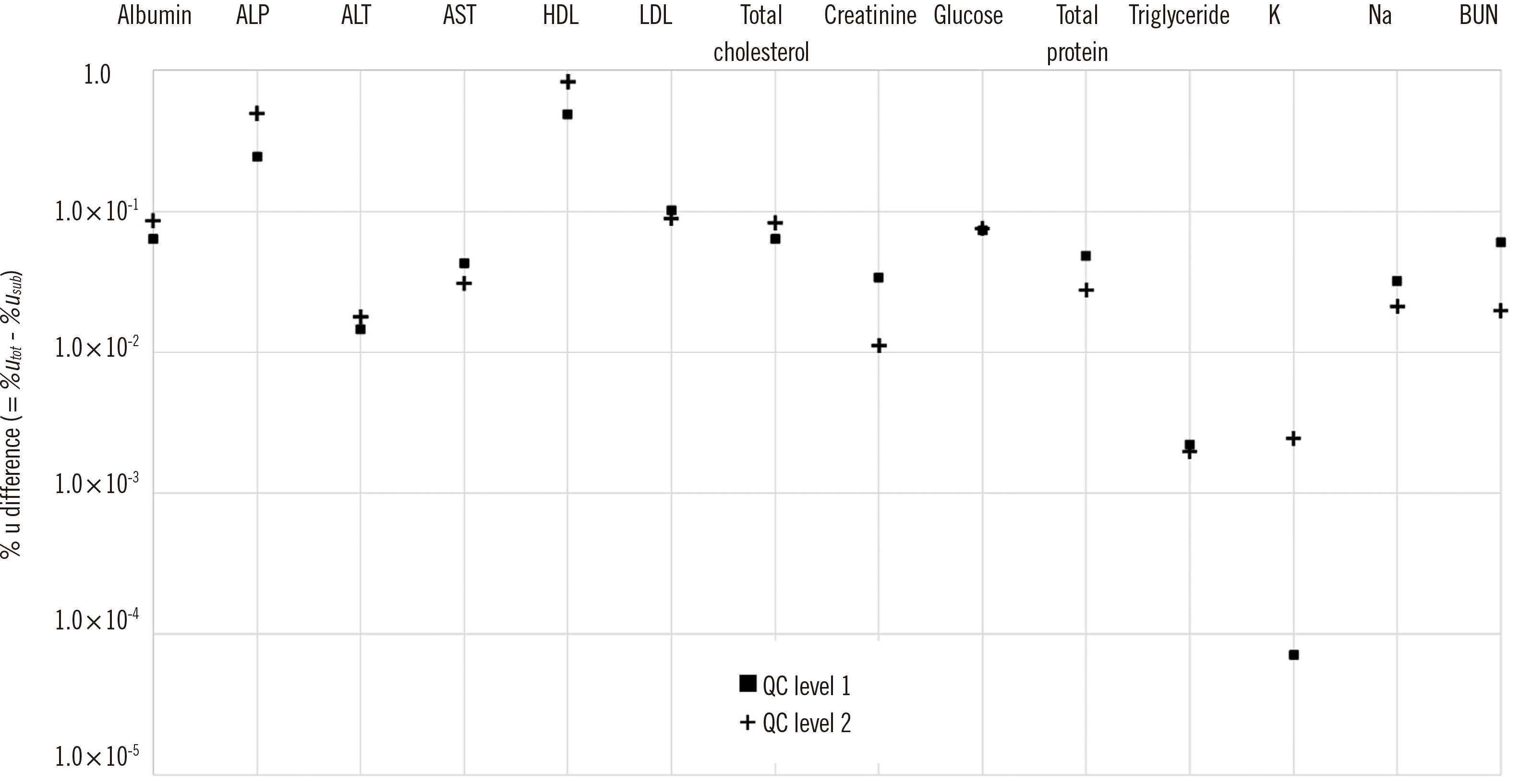
Initially, it was assumed that MU values (%Usub) calculated by subgrouping of the data would be substantially lower than those calculated as a whole (%Utot). However, the differences between MU values obtained by the two different calculation methods were minimal (minimum difference: 7.13×10-5%, maximum difference: 0.825%), although the %Usub values were lower for all analytes.
It is common to observe matrix effects in IQC materials that produce different results than human serum samples during the reaction with reagents [
15]. We attempted to identify how large mean differences between IQC and patient sample results are before and after a reagent lot change (
Supplemental Data Table S2). The mean differences in the two groups were within a narrow interval (in IQC data, up to 3.73% in absolute value; in patient sample data, up to 2.5% in absolute value). We therefore presumed that a mean change of <4% in IQC data may not cause a marked difference, depending on the consideration of a reagent lot change during MU estimation. This may be due to the good management of IQC activities in the laboratory.
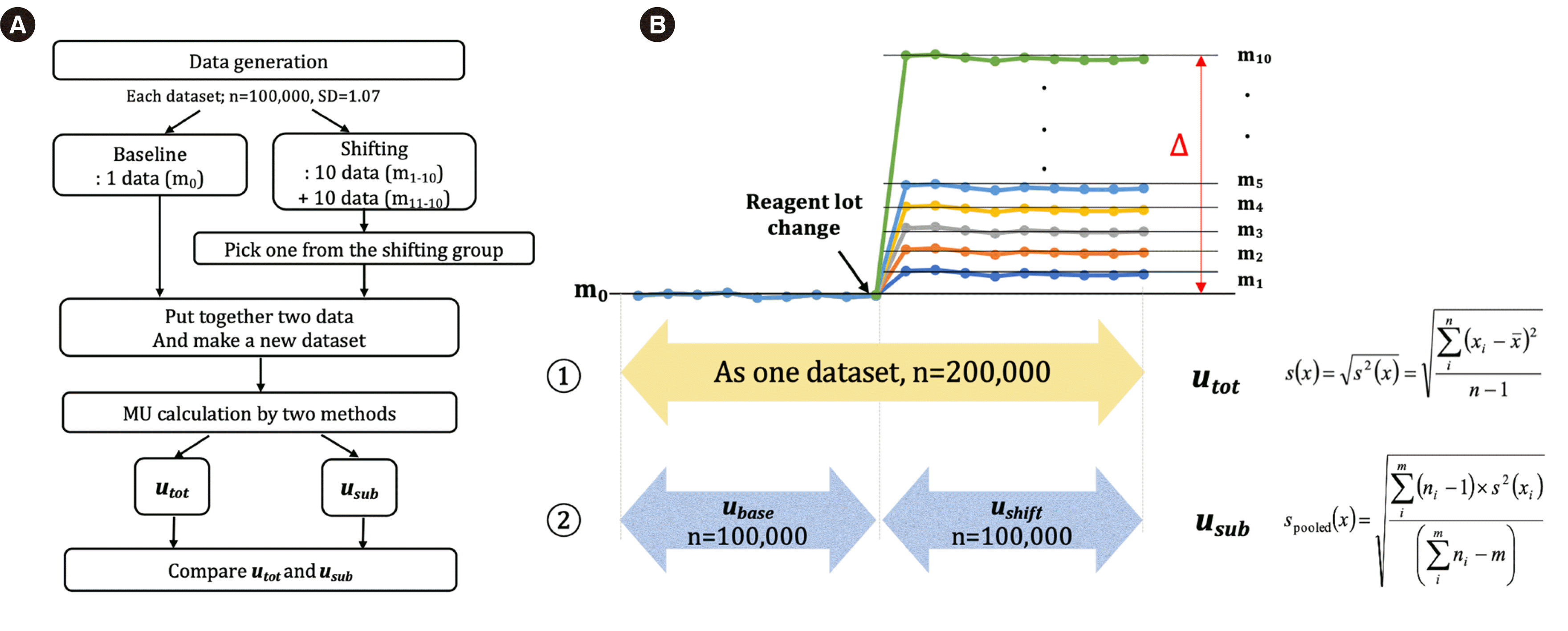
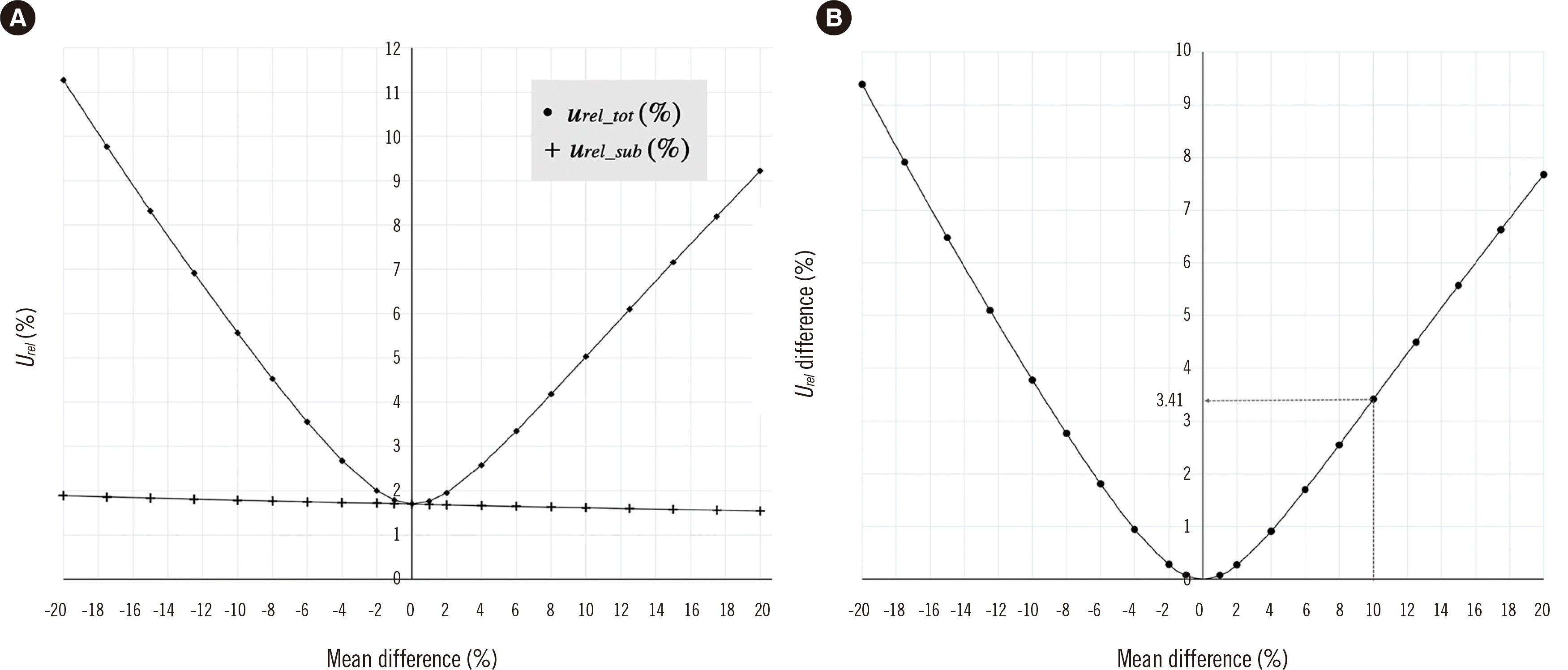
To demonstrate the effect of a significant shift in IQC data after a reagent lot change, we conducted a simulation with artificial IQC datasets consisting of random numbers and considering one reagent lot change. As the degree of IQC data shift gradually increased, the difference between MU results increased according to the calculation method. For example, in a dataset generated with a 10% shift from the mean, the difference in MU values was ~3.41% (
Urel, relative expansion uncertainty,
k=2) (
Fig. 4B). When we comparatively evaluated reagent lot changes in the laboratory, the predefined allowable total error was used as an acceptable performance criterion [
16]. If we presume that the mean difference after a lot change was 8%, which is within the acceptable interval, the new lot would be used without further evaluation. However, in MU estimation, a significant difference was observed depending on the calculation method used. The MU value calculated regardless of the shift of 8% was higher (
utot=4.34) than that calculated considering the shift (
usub=1.7), which led to a highly overestimated MU value.
As observed in the third analysis, a shift in IQC data may indicate that all lots of IQC materials should be treated as different materials. In addition, an SD change in the IQC data may show various uncertainty factors related to the measurement system at the time and/or the IQC material lot change. Therefore, an IQC material lot change may be accompanied by changes not only in the IQC material substances but also in the measurement system over time.
If MU estimation was performed using the combined results of multiple IQC material lot changes, the MU values would be overestimated due to the effects of shifts and other influences from the measurement system over time (6, 7). Therefore, to obtain stable MU values using the TD approach, we suggest that one QC lot should be used for at least six consecutive months.
Lot changes of the calibrator used in MU estimation were not considered in accordance with the ISO guideline, which stipulates that separate collection and calculation of IQC data are not required unless the calibrator manufacturer introduces significant changes, such as a setpoint change [
6,
7]. Furthermore, the calculation using the IQC data as a single set based on the calibrator lot change will capture the variability of human sample results due to this change as a random error [
6,
7]. The means changed in different patterns over several reagent lot changes, which may indicate that the effects of the mean change were weakened. This weakening effect was not considered, and further studies may be needed.
MU estimation in clinical laboratories universally requires more detailed discussions and revisions by expert groups. The results of this study may provide basic, but practical, considerations in clinical laboratories for conducting MU estimation using a TD approach. In conclusion, reagent lot changes should be considered when the TD approach is applied to IQC data, and data from a single lot of IQC materials are recommended to obtain stable and reliable MU values using the TD approach.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download