Abstract
Epithelial-mesenchymal transition (EMT) is known to be involved in airway remodeling and fibrosis of bronchial asthma. However, the molecular mechanisms leading to EMT have yet to be fully clarified. The current study was designed to reveal the potential mechanism of microRNA-21 (miR-21) and poly (ADP-ribose) polymerase-1 (PARP-1) affecting EMT through the PI3K/AKT signaling pathway. Human bronchial epithelial cells (16HBE cells) were transfected with miR-21 mimics/inhibitors and PARP-1 plasmid/small interfering RNA (siRNA). A dual luciferase reporter assay and biotin-labeled RNA pull-down experiments were conducted to verify the targeting relationship between miR-21 mimics and PARP-1. The migration ability of 16HBE cells was evaluated by Transwell assay. Quantitative real-time polymerase chain reaction and Western blotting experiments were applied to determine the expression of Snail, ZEB1, E-cadherin, N-cadherin, Vimentin, and PARP-1. The effects of the PI3K inhibitor LY294002 on the migration of 16HBE cells and EMT were investigated. Overexpression of miR-21 mimics induced migration and EMT of 16HBE cells, which was significantly inhibited by overexpression of PARP-1. Our findings showed that PARP-1 was a direct target of miR-21, and that miR-21 targeted PARP-1 to promote migration and EMT of 16HBE cells through the PI3K/AKT signaling pathway. Using LY294002 to block PI3K/AKT signaling pathway resulted in a significant reduction in the migration and EMT of 16HBE cells. These results suggest that miR-21 promotes EMT and migration of HBE cells by targeting PARP-1. Additionally, the PI3K/AKT signaling pathway might be involved in this mechanism, which could indicate its usefulness as a therapeutic target for asthma.
Asthma is a heterogeneous chronic airway disease characterized by airway inflammation, hyperresponsiveness, and remodeling. The process of airway remodeling is influenced by various inflammatory cells, cytokines, and inflammatory mediators [1]. Recently, it has been found that airway remodeling is positively correlated with the severity of asthma [2]. Airway remodeling mainly includes airway smooth muscle thickening, collagen deposition, and subepithelial fibrosis [3]. At present, there is no effective therapeutic method to reverse airway remodeling. Therefore, to design effective treatment, it is particularly important to understand the exact mechanism between asthma and airway remodeling.
Epithelial-mesenchymal transition (EMT) is a process by which epithelial cells lose their function and characteristics and become mesenchymal cells [4]. During EMT progression, the expression of epithelial markers such as E-cadherin decreases, but that of mesenchymal markers such as Vimentin and N-cadherin increases, and the tight junctions between epithelial cells disappear. The EMT progression changes the function and conformation of epithelial cells into mesenchymal cells [5]. EMT can be categorized into three types according to functional difference: type I is associated with the formation of tissues and organs during the embryonic period, type II is predominantly related to tissue fibrosis, and type III is mainly involved in tumor cell proliferation, metastasis, and invasion. Type II EMT is thought to be associated with airway remodeling. Chronic airway inflammation results in uncontrolled tissue repair through inducing EMT [6]. EMT is also a source of fibroblasts, which play a key role in airway remodeling [7]. During chronic airway inflammation, EMT promotes the migration of epithelial cells and excessive secretion of extracellular matrix to narrow or block the bronchus, which may eventually worsen asthma [8].
MicroRNAs (miRs) are noncoding, single-stranded RNAs that regulate gene expression at the post-transcriptional level [9]. MiRs base-pair with target messenger RNAs (mRNAs) to silence complexes and degrade mRNAs or block translation, thus influencing protein synthesis [10]. About 30% of the human genome is targeted by miRs that repress translation or degrade target genes [11], playing an important role in lung diseases including cancer, asthma, and chronic obstructive pulmonary disease. It has been reported that miR-1, miR-21, miR-126, and miR-221 are involved in the progression of asthma [12]. MiR-21 could reduce the expression level of phosphatase and tensin homolog deleted on chromosome 10 (PTEN) by activating the PI3K/AKT signaling pathway, which promotes the proliferation and migration of human airway smooth muscle cells [13]. Many studies have suggested that miR-21 is associated with asthma and is a potential therapeutic target for asthma. However, the precise mechanism of miR-21 in asthma needs to be further explored.
Poly (ADP-ribose) polymerases (PARPs) are a kind of DNA repair enzyme that play an important role in cell repair and apoptosis [14]. Among all protein family members, PARP-1 is reportedly significant in allergen-induced inflammation and airway hyper-responsiveness [15]. Additionally, PARP-1 could promote airway remodeling through inducing nitric oxide and transforming growth factor-β (TGF-β) synthesis [16]. Research has shown that use of a selective PARP-1 inhibitor reduced allergen-induced asthma-like reaction, bronchial hyper-responsiveness, and airway remodeling [17]. TGF-β has been described as one of the main inducers of EMT and activators of the PI3K/AKT signaling pathway [18,19]. Inhibiting the PI3K/AKT signaling pathway alleviated lung inflammation by reducing the expression of interleukin (IL)-4, IL-6, IL-8, tumor necrosis factor-α, and immunoglobulin E. In addition, inhibition of the PI3K/AKT signaling pathway could suppress proliferation of airway smooth muscle cells and airway remodeling. We previously used high-throughput sequencing and bioinformatics analysis to demonstrate that PARP-1, as a downstream target gene of miR-21, might participate in the occurrence and development of combined allergic rhinitis and asthma syndrome (CARAS) [20].
The aim of the present study was to evaluate the role of miR-21/PARP-1 in the EMT and migration of human bronchial epithelial (16HBE) cells, focusing on the relationship between miR-21/PARP-1 and the PI3K/AKT molecular signaling pathway.
Human bronchial epithelial (16HBE) cells were purchased from Zhongqiaoxinzhou Biotech (Shanghai, China). All cells were cultured in Dulbecco’s modified Eagle medium (DMEM) (Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum, penicillin, and streptomycin (Thermo Fisher Scientific, Carlsbad, CA, USA) at 37°C with humidified 5% CO2. Once the cells reached 70%–80% confluence, the medium was replaced with DMEM containing 1% fetal bovine serum and 30 μM PI3K inhibitor LY294002 (MedChemExpress, New Jersey, NJ, USA) dissolved in dimethylsulfoxide (Sigma, St. Louis, MI, USA) for 24 h. The cells were incubated with 0.6% of dimethylsulfoxide as a normal control. The concentration of dimethylsulfoxide was the same in both normal control and experiment groups.
The hsa-miR-21 mimics/inhibitor and PARP-1 plasmid/small interfering RNA (siRNA) were obtained from GenePharma company (Suzhou, China). Cells were seeded onto 6-well plates until reaching 60%–70% confluence. According to the manufacturer’s instructions, cells were transfected with hsa-miR-21 mimics/inhibitor (10 nM) and PARP-1 plasmid/siRNA (3 μg) using Lipofectamine 2000 (Thermo Fisher Scientific, Boston, MA, USA). Cells were harvested and extracted for further study 48 h post-transfection.
The migration of 16HBE cells was evaluated by Transwell assay with 8 μm Transwell filters. Cells transfected with hsa-miR-21 mimics/hsa-miR-21 inhibitor, PARP-1 plasmid/PARP-1 siRNA, and controls were incubated in the upper chamber with 100 μl DMEM without fetal bovine serum at a density of 3 × 104 cells/well for 24 h. The migration of 16HBE cells was stimulated by adding DMEM with 10% fetal bovine serum into the lower chamber. Finally, the cells that migrated through the membrane were fixed in 4% paraformaldehyde and stained with Giemsa. The migration ability of cells was observed and photographed using an inverted phase contrast microscope. The experiments were replicated three times.
Total RNA from the 16HBE cells was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The first strand of complementary DNA was synthesized by the First Strand cDNA Synthesis Kit (Vazyme, Nanjing, China) following the manufacturer’s instructions. Then, qRT-PCR (ABI PRISM7500; ABI, Foster City, CA, USA) was performed to analyze the expression levels of RNA with AceQ qPCR SYBR Green Master Mix and miRNA Universal SYBR qPCR Master Mix (Vazyme). The specific primers used for qPCR were designed and synthesized by GenePharma Company (Table 1). β-Actin was regarded as a housekeeping gene for E-cadherin, N-cadherin, Vimentin, ZEB1, Snail, and PARP-1, while U6 was used as inner control for miR-21 mimics. The ∆∆Ct method was used for estimating the gene expression levels. All experiments were performed three times.
Protein samples from 16HBE cells were extracted with RIPA lysis buffer (Beyotime, Shanghai, China) supplemented with protease inhibitors and phosphatase inhibitor (Beyotime) at 4°C for 30 min. Then, the concentration of protein was calculated using the BCA Protein Assay kit (Beyotime). Protein samples were subjected to 8% sodium dodecyl sulfate polyacrylamide gel electrophoresis and electroblotted onto polyvinylidene membrane. Subsequently, the membranes were blocked with 5% skimmed milk. The primary antibodies against E-cadherin, N-cadherin, Vimentin, PI3K, p-AKT, t-AKT, PARP-1 (CST, Boston, MA, USA), Snail, ZEB1 (ABclonal, Wuhan, China), and GAPDH (Abcam, Cambridge, UK) were incubated with the membranes at 4°C overnight. Then the membranes were washed three times and incubated for 2 h with horseradish peroxidase-conjugated goat antirabbit Immunoglobulin G secondary antibody at room temperature (Abcam). Finally, the protein on the membrane was visualized with the Chemiluminescent ECL Detection System (Tanon, Shanghai, China) and quantitated by ImageJ version 1.48 (National Institutes of Health, Bethesda, MD, USA).
Wild and mutant 3’-UTRs of PARP-1 chemically synthesised were cloned into the pmirGLO dual-luciferase miRNA target expression vector (Promega Corp., Madison, WI, USA) between the PmeI and XbaI sites. Both sense and antisense primers of wild and mutant PARP-1 which had an internal NotI restriction site. The wild and mutant clones containing the insertion sequence of PARP-1 3’UTR were tested using the NotI restriction enzyme. MiR-21 mimics or negative control miRNA were co-transfected with wild or mutant PARP-1 3’UTR constructs (200 ng) using Lipofectamine 2000 transfection reagent (Invitrogen). Passive lysis buffer was used to prepare cell lysate after 24 h of transfection, and the luciferase activity was recorded using the dual luciferase reporter assay method (Promega Corp.) as instructed by manufacturers. Firefly activity was normalized to Renilla activity. The experiments were performed three times.
T4 RNA ligase was attached to a single biotinylated nucleotide to the 3’ terminus of miR-21 strands and normal controls to synthesize the biotin-labeled miR-21 mimics probes and normal control probes according to the manufacturer’s instructions of the Pierce RNA 3’ End Desthiobiotinylation Kit (Thermo Fisher Scientific). Then we used the biotin-labeled RNA probes to bind Streptavidin Magnetic Beads (Thermo Fisher Scientific) for 1 h at room temperature. Whole cell lysates were incubated with the mixture RNA-breads overnight at 4°C, the leftover whole cell lysates as the input group. Finally, the RNA-protein complexes and input group were detected by Western blot.
All experimental data were from three independent replicates and expressed as mean ± standard deviation (SD). GraphPad Prism software version 8.0 (GraphPad Software, San Diego, CA, USA) was used to analyze all data. Differences between the groups were assessed by one-way analysis of variance or Student’s paired t-test, as appropriate. A p-value of < 0.05 was considered statistically significant.
The 16HBE cells were transfected with PARP-1 plasmid or PARP-1 siRNA to explore the effect on migration ability. The protein and mRNA expression levels of PARP-1 increased in 16HBE cells transfected with PARP-1 plasmid, and decreased in 16HBE cells transfected with PARP-1 siRNA (Fig. 1A, B). The Transwell assay revealed that PARP-1 significantly inhibited the migration ability of 16HBE cells (Fig. 1C). Furthermore, blocking PARP-1 with PARP-1 siRNA increased the number of migrated 16HBE cells (Fig. 1C). Therefore, PARP-1 suppressed the migration of 16HBE cells.
As shown in Fig. 2A, the transfection of miR-21 mimics and miR-21 inhibitor into 16HBE cells was successful. Fig. 2B and C show the effects of miR-21 mimics on PARP-1 protein and mRNA. The protein and mRNA expressions of PARP-1 were inhibited significantly by miR-21 mimics. In contrast, the protein and mRNA expression levels of PARP-1 increased in cells transfected with miR-21 inhibitor. However, the changes in PARP-1 induced by miR-21 mimics or miR-21 inhibitor were reversed by PARP-1 plasmid or PARP-1 siRNA. As bioinformatics analysis showed, PARP-1 was predicted as a potential target of miR-21 (Fig. 2D). We verified the target-regulatory relationship between miR-21 and PARP-1 by dual-luciferase reporter assay. As Fig. 2D illustrates, miR-21 mimics could bind to PARP-1 mRNA and promote its degradation in the wild-type 3’UTR PARP-1 group, while they did not affect the stability of PARP-1 expression in the mutant 3’UTR PARP-1 group. Western blot with anti-PARP-1 antibody detected the existence of PARP-1 within biotin-labeled miR-21 mimics probes pull-down samples, but the normal control probes group did not detect the existence of PARP-1 (Fig. 2E). These findings indicate that miR-21 targets PARP-1 directly.
As shown in Fig. 2A, PARP-1 is a direct target of miR-21. The cells were transfected with miR-21 mimics or miR-21 inhibitor to study the effects of miR-21 mimics on the migration ability of 16HBE cells. Fig. 3A illustrates that the migration ability of 16HBE cells was enhanced with overexpression of miR-21 mimics, but reduced with cotransfection of PARP-1 plasmid. The opposite results were observed in 16HBE cells transfected with miR-21 inhibitor, and reversed by co-transfecting PARP-1 siRNA (Fig. 3B). Phase contrast microscope images of 16HBE cells transfected with normal control, miR-21 mimics, miR-21 inhibitor, PARP-1 plasmid, or PARP-1 siRNA show that 16HBE cells transfected with miR-21 mimics or PARP-1 siRNA adopted a spindle-like shape, whereas cells transfected with normal control, miR-21 inhibitor, or PARP-1 plasmid adopted a cobblestone-like appearance (Fig. 3C) These results indicate that miR-21 could promote the migration of 16HBE cells via regulating the expression of PARP-1 and further implicate the influence of miR-21 mimics and PARP-1 on EMT in 16HBE cells.
To elucidate the effect of PARP-1 on the EMT of 16HBE cells, we used Western blot analysis and qRT-PCR to evaluate the expression of EMT-regulating transcription factors and EMT-related proteins. The Western blot analysis demonstrated that PARP-1 significantly increased the protein expression of the epithelial marker E-cadherin, but decreased the protein expression of mesenchymal markers Vimentin and N-cadherin, as well as EMT-regulating transcription factors Snail and ZEB1. Meanwhile, the opposite results were observed in cells transfected with PARP-1 siRNA (Fig. 4A, C). Fig. 4B and D illustrate consistent changes in mRNA expression levels of EMT-regulating transcription factors and EMT-related proteins. These results suggest that PARP-1 suppresses the EMT of 16HBE cells.
To identify the role of the miR-21/PARP-1 axis on the EMT of 16HBE cells, we transfected normal controls, miR-21 mimics, miR-21 inhibitor, miR-21 mimics + PARP-1 plasmid, or miR-21 inhibitor + siPARP-1 into 16HBE cells. As presented in Fig. 5A, B, E, and F, miR-21 mimics could increase the protein and mRNA expressions of EMT-regulating transcription factors Snail and ZEB1 and mesenchymal marker N-cadherin and Vimentin, but decrease the protein and mRNA expression of epithelial marker E-cadherin. Further, the changes of EMT-related proteins N-cadherin, Vimentin, and E-cadherin could be reversed by PARP-1 overexpression (Fig. 5E, F). Opposite results were observed in 16HBE cells transfected with miR-21 inhibitor or miR-21 inhibitor + siPARP-1 (Fig. 5C, D, G, and H). The results suggest that overexpression of PARP-1 could alleviate the EMT of 16HBE cells induced by miR-21 mimics.
To further evaluate a possible molecular mechanism involved in the EMT of 16HBE cells induced by miR-21/PARP-1, we used Western blot analysis to detect molecular proteins related to the PI3K/AKT signaling pathway. The results revealed that PARP-1 reduced the protein expressions of p-AKT and PI3K (Fig. 6A). MiR-21 mimics promoted the protein expressions of p-AKT and PI3K, and the elevated proteins of p-AKT and PI3K promoted by miR-21 mimics could be reversed by PARP-1 overexpression. MiR-21 inhibitor played an opposite role in the protein expressions of p-AKT and PI3K. Also, the effect of miR-21 inhibitor on the protein expressions of p-AKT and PI3K was reversed by PARP-1 siRNA (Fig. 6C). However, as indicated in Fig. 6A–C, there were no changes in the protein expression level of t-AKT in each group. In summary, miR-21 activated the PI3K/AKT signaling pathway by targeting PARP-1.
To further clarify the effect of the PI3K/AKT signaling pathway on regulating the migration of 16HBE cells, we co-cultured the 16HBE cells with LY294002, a PI3K inhibitor, under the starvation condition before transfection. The results showed that LY294002 could reduce the number of migrated 16HBE cells induced by miR-21 mimics (Fig. 7A). Identical effects of LY294002 on migration ability were found in 16HBE cells transfected with PARP-1 siRNA (Fig. 7B). These findings indicate that miR-21/PARP-1 might regulate the migration of 16HBE cells through the PI3K/AKT signaling pathway.
We administered the PI3K inhibitor LY294002 to 16HBE cells before transfecting them with miR-21 mimics or PARP-1 siRNA to determine the effect on the PI3K/AKT signaling pathway. As illustrated in Figs. 8A and B, activation of the pathway by miR-21 mimics or PARP-1 siRNA was blocked by LY294002. LY294002 also remarkably ameliorated the decreased protein expression of epithelial marker E-cadherin and increased protein expression of mesenchymal markers Vimentin and N-cadherin induced by miR-21 mimics or PARP-1 siRNA. However, LY294002 had no effect on the protein expression of PARP-1 (Fig. 8C, E). Comparable results from qRT-PCR were also observed (Fig. 8D, F). Thus, these findings demonstrate that miR-21/PARP-1 induces the EMT and migration of 16HBE cells via activation of the PI3K/AKT signaling pathway, as illustrated in Fig. 9.
Several previous studies have reported on the role of miR-21 and PARP-1 in the development of asthma [12,13,18]. Our previous research, targeting PARP-1 using high-throughput sequencing and bioinformatics analysis, reported that miR-21 might be involved in the pathogenesis of CARAS [20]. We also found that miR-21/PARP-1 regulated the apoptosis and proliferation of 16HBE cells in vitro (data unpublished). However, no data are currently available on the role of miR-21 in the pathological mechanism of asthma via regulating PARP-1. The present study analyzed the expression levels of EMT-related proteins and EMT-regulating transcription factors and changes of migration ability to explore the effects of miR-21/PARP-1 on the EMT in 16HBE cells. The results suggested that miR-21 induced the EMT and migration of 16HBE cells by targeting PARP-1, and the PI3K/AKT signaling pathway likely participated in the molecular mechanism.
P110γ and P110δ are catalytic subunits of PI3K, which are dramatically expressed in lymphocytes and related to neutrophil activation and mast cell degranulation, eventually contributing to the development of inflammation. PTEN could downregulate PI3K and maintain normal immune balance to avoid immune-related disease [21]. Many studies have suggested that the PI3K/AKT signaling pathway plays an important role in the pathogenesis of asthma and that inhibition of this pathway could alleviate the inflammation of lung disease. For example, activation of the TLR4-PI3Kδ axis was related to nuclear translocation of nuclear factor-κB and mitochondrial reactive oxygen species generation in house-dust-mite-induced allergic asthma [22]. In allergic asthma, IL-17A activated the PI3K/AKT signaling pathway and further promoted the secretion of IL-8 and TGF-β1, which resulted in EMT and exacerbated asthma [23,24].
MiR21 has been reported to be highly expressed as an oncogene in various human cancers, including lung, breast, ovarian, colon, rectal, and prostate cancer [25]. Analysis of lung cancer biopsy specimens showed that miR-21-5p was increased significantly in lung adenocarcinoma compared with adjacent tissues and cells [26]. The levels of plasma miR-21 in asthmatic children were upregulated, and miR-21 was a better asthma biomarker than other miRs [10]. Analysis of transcriptional regulation of the gene expression signature also suggested that the levels of miR-21 in people with CARAS were higher than those in a control group [20]. Previous research confirmed that TGF-β1 stimulated proliferation of airway smooth muscle cells in rats via Smad2/3-signaling-pathway-mediated upregulation of miR-21 and phosphorylation of the PI3K/AKT signaling pathway [27]. Moreover, in vivo study found that infection-induced miR-21 expression promoted phosphorylation of PI3K/AKT, which suppressed histone deacetylase 2 and resulted in steroid insensitivity [28].
It has been reported that LncRNA-CASC7 enhanced the corticosteroid sensitivity of airway smooth muscle cells via inhibiting the PI3K/AKT signaling pathway by targeting miR-21 [29]. In addition, inhibiting the expression of miR-21 has been shown to upregulate Smad7 protein expression, which could attenuate the airway remodeling aggravated by the TGF-β/Smad signaling pathway [30] and reverse the EMT of pulmonary epithelial cells induced by TGF-β [31]. Furthermore, inhibiting miR-21 expression could reduce bleomycin-induced pulmonary fibrosis in rats [32]. MiR-21 is overexpressed in non-small-cell lung cancer compared to adenocarcinomas, squamous cell carcinomas, and these mixed tumors; Dai et al. [33] did research on the five types of human lung cancer cells and found that miR-21 promoted EMT via PTEN/AKT/GSK3β signaling, which regulates the proliferation, migration and apoptosis of lung cancer cells. Our study found that the PI3K inhibitor LY294002 could inhibit the EMT and migration induced by miR-21 in 16HBE cells in vitro. Our findings consistently suggested that miR-21 works as an inducer of EMT by promoting the PI3K/AKT signaling pathway in 16HBE cells.
The activation of PARP-1 could post-translationally modify transcription factors by poly (ADP-ribosylation) which affects the transcription of inflammation genes. Recent in vivo studies suggest that activation of PARP-1 contributes to airway inflammation in numerous animal models. In one study, inhibition of PARP-1 prevented eosinophil recruitment via modulating Th2 cytokines, thus alleviating allergic airway inflammation in a murine model [34]. Other research found that activation of PARP-1 was critical for migration of dendritic cells from peripheral blood to the lung in a murine model, but not associated with differentiation and function of dendritic cells [35]. Another study showed that PARP was activated in peripheral blood and lung tissue in asthmatic patients, and inhibition of PARP by gene knockout or Olaparib could block established asthmatic features, including production of Th2 cytokines and mucus, and airway hyper-responsiveness [36]. All of these studies showed a protective effect of PARP-1 inhibition against asthma. Conversely, our study demonstrated that PARP-1 is a direct target of miR-21 and is negatively regulated by it. Silencing PARP-1 could induce EMT, as well as promote the migration of 16HBE cells by activating the PI3K/AKT signaling pathway. The contradiction may be explained by the different studies being in vitro versus in vivo. Thus, further study is needed to ascertain the biological function of PARP-1.
Severe asthma is defined as that which requires a high dose of inhaled corticosteroids plus a second controller or systemic corticosteroids to prevent it from becoming “uncontrolled,” or asthma that remains ‘‘uncontrolled’’ despite this therapy [37]. There has been evidence that recurring EMT could lead to airway remodeling and eventually exacerbate asthma. EMT has been considered a gradual process and a reversible condition. However, recent research suggests that airway remodeling happens in the early onset of asthma, but not after airway inflammation [38]. Therefore, inhibition of EMT could reverse the airway remodeling and prevent the exacerbation of asthma. Our study indicates that miR-21 and PARP-1 may be potential therapeutic targets for severe asthma.
In summary, our study showed that miR-21 could promote the EMT and migration of HBE cells by targeting and downregulating PARP-1 and activating the PI3K/AKT signaling pathway. Further study on the interaction of miR-21, PARP-1, and the PI3K/AKT signaling pathway in asthma is necessary and promising.
Notes
REFERENCES
1. De Ferrari L, Chiappori A, Bagnasco D, Riccio AM, Passalacqua G, Canonica GW. 2016; Molecular phenotyping and biomarker development: are we on our way towards targeted therapy for severe asthma? Expert Rev Respir Med. 10:29–38. DOI: 10.1586/17476348.2016.1111763. PMID: 26566089. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84946866022&origin=inward.

2. Loffredo LF, Abdala-Valencia H, Anekalla KR, Cuervo-Pardo L, Gottardi CJ, Berdnikovs S. 2017; Beyond epithelial-to-mesenchymal transition: common suppression of differentiation programs underlies epithelial barrier dysfunction in mild, moderate, and severe asthma. Allergy. 72:1988–2004. DOI: 10.1111/all.13222. PMID: 28599074. PMCID: PMC5698119. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85021803995&origin=inward.

3. Jeffery PK. 2001; Remodeling in asthma and chronic obstructive lung disease. Am J Respir Crit Care Med. 164(10 Pt 2):S28–S38. DOI: 10.1164/ajrccm.164.supplement_2.2106061. PMID: 11734464. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=0035891664&origin=inward.
4. Larriba MJ, García de Herreros A, Muñoz A. 2016; Vitamin D and the epithelial to mesenchymal transition. Stem Cells Int. 2016:6213872. DOI: 10.1155/2016/6213872. PMID: 26880977. PMCID: PMC4736588. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84961353431&origin=inward.

5. Xia H, Xue J, Xu H, Lin M, Shi M, Sun Q, Xiao T, Dai X, Wu L, Li J, Xiang Q, Tang H, Bian Q, Liu Q. 2019; Andrographolide antagonizes the cigarette smoke-induced epithelial-mesenchymal transition and pulmonary dysfunction through anti-inflammatory inhibiting HOTAIR. Toxicology. 422:84–94. DOI: 10.1016/j.tox.2019.05.009. PMID: 31128153. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85066128653&origin=inward.

6. Haddad A, Gaudet M, Plesa M, Allakhverdi Z, Mogas AK, Audusseau S, Baglole CJ, Eidelman DH, Olivenstein R, Ludwig MS, Hamid Q. 2019; Neutrophils from severe asthmatic patients induce epithelial to mesenchymal transition in healthy bronchial epithelial cells. Respir Res. 20:234. DOI: 10.1186/s12931-019-1186-8. PMID: 31665016. PMCID: PMC6819645. PMID: f8111fbb356345d28571017c5e74b51b. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85074433350&origin=inward.

7. Hackett TL. 2012; Epithelial-mesenchymal transition in the pathophysiology of airway remodelling in asthma. Curr Opin Allergy Clin Immunol. 12:53–59. DOI: 10.1097/ACI.0b013e32834ec6eb. PMID: 22217512. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84856896936&origin=inward.

8. Pain M, Bermudez O, Lacoste P, Royer PJ, Botturi K, Tissot A, Brouard S, Eickelberg O, Magnan A. 2014; Tissue remodelling in chronic bronchial diseases: from the epithelial to mesenchymal phenotype. Eur Respir Rev. 23:118–130. DOI: 10.1183/09059180.00004413. PMID: 24591669.

9. Moret-Tatay I, Iborra M, Cerrillo E, Tortosa L, Nos P, Beltrán B. 2016; Possible biomarkers in blood for Crohn's disease: oxidative stress and microRNAs-current evidences and further aspects to unravel. Oxid Med Cell Longev. 2016:2325162. DOI: 10.1155/2016/2325162. PMID: 26823944. PMCID: PMC4707323. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84957061022&origin=inward.

10. Hammad Mahmoud Hammad R, Hamed DHED, Eldosoky MAER, Ahmad AAES, Osman HM, Abd Elgalil HM, Mahmoud Hassan MM. 2018; Plasma microRNA-21, microRNA-146a and IL-13 expression in asthmatic children. Innate Immun. 24:171–179. DOI: 10.1177/1753425918763521. PMID: 29635981. PMCID: PMC6852388. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85045521025&origin=inward.

11. Gurtan AM, Sharp PA. 2013; The role of miRNAs in regulating gene expression networks. J Mol Biol. 425:3582–3600. DOI: 10.1016/j.jmb.2013.03.007. PMID: 23500488. PMCID: PMC3757117. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84883805211&origin=inward.

12. Rebane A, Akdis CA. 2014; MicroRNAs in allergy and asthma. Curr Allergy Asthma Rep. 14:424. DOI: 10.1007/s11882-014-0424-x. PMID: 24504527. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84893156260&origin=inward.

13. Liu Y, Yang K, Shi H, Xu J, Zhang D, Wu Y, Zhou S, Sun X. 2015; MiR-21 modulates human airway smooth muscle cell proliferation and migration in asthma through regulation of PTEN expression. Exp Lung Res. 41:535–545. DOI: 10.3109/01902148.2015.1090501. PMID: 26651881. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84949768176&origin=inward.

14. Krishnakumar R, Kraus WL. 2010; The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol Cell. 39:8–24. DOI: 10.1016/j.molcel.2010.06.017. PMID: 20603072. PMCID: PMC2923840. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=77954274504&origin=inward.

15. Naura AS, Hans CP, Zerfaoui M, You D, Cormier SA, Oumouna M, Boulares AH. 2008; Post-allergen challenge inhibition of poly(ADP-ribose) polymerase harbors therapeutic potential for treatment of allergic airway inflammation. Clin Exp Allergy. 38:839–846. DOI: 10.1111/j.1365-2222.2008.02943.x. PMID: 18261157. PMCID: PMC2740645. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=42249103606&origin=inward.

16. Naura AS, Zerfaoui M, Kim H, Abd Elmageed ZY, Rodriguez PC, Hans CP, Ju J, Errami Y, Park J, Ochoa AC, Boulares AH. 2010; Requirement for inducible nitric oxide synthase in chronic allergen exposure-induced pulmonary fibrosis but not inflammation. J Immunol. 185:3076–3085. DOI: 10.4049/jimmunol.0904214. PMID: 20668217. PMCID: PMC3077076. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=78049413973&origin=inward.

17. Lucarini L, Pini A, Gerace E, Pellicciari R, Masini E, Moroni F. 2014; Poly(ADP-ribose) polymerase inhibition with HYDAMTIQ reduces allergen-induced asthma-like reaction, bronchial hyper-reactivity and airway remodelling. J Cell Mol Med. 18:468–479. DOI: 10.1111/jcmm.12197. PMID: 24444146. PMCID: PMC3955153. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84894567125&origin=inward.

18. Kim KK, Kugler MC, Wolters PJ, Robillard L, Galvez MG, Brumwell AN, Sheppard D, Chapman HA. 2006; Alveolar epithelial cell mesenchymal transition develops in vivo during pulmonary fibrosis and is regulated by the extracellular matrix. Proc Natl Acad Sci U S A. 103:13180–13185. DOI: 10.1073/pnas.0605669103. PMID: 16924102. PMCID: PMC1551904. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=33748375828&origin=inward.

19. Hamidi A, Song J, Thakur N, Itoh S, Marcusson A, Bergh A, Heldin CH, Landström M. 2017; TGF-β promotes PI3K-AKT signaling and prostate cancer cell migration through the TRAF6-mediated ubiquitylation of p85α. Sci Signal. 10:eaal4186. DOI: 10.1126/scisignal.aal4186. PMID: 28676490. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85022054230&origin=inward.

20. Mao Z, Shi Y, Cao Q, Chen Y, Sun Y, Liu Z, Zhang Q, Huang M. 2018; Transcriptional regulation on the gene expression signature in combined allergic rhinitis and asthma syndrome. Epigenomics. 10:119–131. DOI: 10.2217/epi-2017-0072. PMID: 29334241. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85041135177&origin=inward.

21. Suzuki A, Yamaguchi MT, Ohteki T, Sasaki T, Kaisho T, Kimura Y, Yoshida R, Wakeham A, Higuchi T, Fukumoto M, Tsubata T, Ohashi PS, Koyasu S, Penninger JM, Nakano T, Mak TW. 2001; T cell-specific loss of Pten leads to defects in central and peripheral tolerance. Immunity. 14:523–534. DOI: 10.1016/S1074-7613(01)00134-0. PMID: 11371355. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=0034995242&origin=inward.

22. Kim SR, Park HJ, Lee KB, Kim HJ, Jeong JS, Cho SH, Lee YC. 2020; Epithelial PI3K-δ promotes house dust mite-induced allergic asthma in NLRP3 inflammasome-dependent and -independent manners. Allergy Asthma Immunol Res. 12:338–358. DOI: 10.4168/aair.2020.12.2.338. PMID: 32009326. PMCID: PMC6997282. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85082684615&origin=inward.

23. Cohen L, E X, Tarsi J, Ramkumar T, Horiuchi TK, Cochran R, DeMartino S, Schechtman KB, Hussain I, Holtzman MJ, Castro M. 2007; Epithelial cell proliferation contributes to airway remodeling in severe asthma. Am J Respir Crit Care Med. 176:138–145. DOI: 10.1164/rccm.200607-1062OC. PMID: 17463414. PMCID: PMC1994213. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=34447318645&origin=inward.

24. Liu G, Zhu R, Li B. 2005; TNF-alpha and IL-8 of the patients with allergic asthma. J Huazhong Univ Sci Technolog Med Sci. 25:274–275. 309DOI: 10.1007/BF02828140. PMID: 16201269.
25. Pfeffer SR, Yang CH, Pfeffer LM. 2015; The role of miR-21 in cancer. Drug Dev Res. 76:270–277. DOI: 10.1002/ddr.21257. PMID: 26082192. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84941259771&origin=inward.

26. Wang G, Zhou Y, Chen W, Yang Y, Ye J, Ou H, Wu H. 2020; miR-21-5p promotes lung adenocarcinoma cell proliferation, migration and invasion via targeting WWC2. Cancer Biomark. 28:549–559. DOI: 10.3233/CBM-201489. PMID: 32623387. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85089709669&origin=inward.

27. Liu L, Pan Y, Zhai C, Zhu Y, Ke R, Shi W, Wang J, Yan X, Su X, Song Y, Gao L, Li M. 2018; Activation of peroxisome proliferation-activated receptor-γ inhibits transforming growth factor-β1-induced airway smooth muscle cell proliferation by suppressing Smad-miR-21 signaling. J Cell Physiol. 234:669–681. DOI: 10.1002/jcp.26839. PMID: 30132829. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85052377345&origin=inward.

28. Kim RY, Horvat JC, Pinkerton JW, Starkey MR, Essilfie AT, Mayall JR, Nair PM, Hansbro NG, Jones B, Haw TJ, Sunkara KP, Nguyen TH, Jarnicki AG, Keely S, Mattes J, Adcock IM, Foster PS, Hansbro PM. 2017; MicroRNA-21 drives severe, steroid-insensitive experimental asthma by amplifying phosphoinositide 3-kinase-mediated suppression of histone deacetylase 2. J Allergy Clin Immunol. 139:519–532. DOI: 10.1016/j.jaci.2016.04.038. PMID: 27448447. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84979590841&origin=inward.

29. Liu JH, Li C, Zhang CH, Zhang ZH. 2020; LncRNA-CASC7 enhances corticosteroid sensitivity via inhibiting the PI3K/AKT signaling pathway by targeting miR-21 in severe asthma. Pulmonology. 26:18–26. DOI: 10.1016/j.pulmoe.2019.07.001. PMID: 31412983. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85070301507&origin=inward.

30. Yu ZW, Xu YQ, Zhang XJ, Pan JR, Xiang HX, Gu XH, Ji SB, Qian J. 2019; Mutual regulation between miR-21 and the TGFβ/Smad signaling pathway in human bronchial fibroblasts promotes airway remodeling. J Asthma. 56:341–349. DOI: 10.1080/02770903.2018.1455859. PMID: 29621415. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85045072483&origin=inward.

31. Yamada M, Kubo H, Ota C, Takahashi T, Tando Y, Suzuki T, Fujino N, Makiguchi T, Takagi K, Suzuki T, Ichinose M. 2013; The increase of microRNA-21 during lung fibrosis and its contribution to epithelial-mesenchymal transition in pulmonary epithelial cells. Respir Res. 14:95. DOI: 10.1186/1465-9921-14-95. PMID: 24063588. PMCID: PMC3849377. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84884494821&origin=inward.

32. Gao Y, Lu J, Zhang Y, Chen Y, Gu Z, Jiang X. 2013; Baicalein attenuates bleomycin-induced pulmonary fibrosis in rats through inhibition of miR-21. Pulm Pharmacol Ther. 26:649–654. DOI: 10.1016/j.pupt.2013.03.006. PMID: 23523661. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84888009848&origin=inward.

33. Dai L, Chen F, Zheng Y, Zhang D, Qian B, Ji H, Long F, Cretoiu D. 2019; miR-21 regulates growth and EMT in lung cancer cells via PTEN/Akt/GSK3β signaling. Front Biosci (Landmark Ed). 24:1426–1439. DOI: 10.2741/4788. PMID: 31136988. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85067277602&origin=inward.
34. Oumouna M, Datta R, Oumouna-Benachour K, Suzuki Y, Hans C, Matthews K, Fallon K, Boulares H. 2006; Poly(ADP-ribose) polymerase-1 inhibition prevents eosinophil recruitment by modulating Th2 cytokines in a murine model of allergic airway inflammation: a potential specific effect on IL-5. J Immunol. 177:6489–6496. Erratum. DOI: 10.4049/jimmunol.177.9.6489. PMID: 17056581. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=33750314513&origin=inward.

35. Echeverri Tirado LC, Ghonim MA, Wang J, Al-Khami AA, Wyczechowska D, Luu HH, Kim H, Sanchez-Pino MD, Yélamos J, Yassin LM, Boulares AH. 2019; PARP-1 is critical for recruitment of dendritic cells to the lung in a mouse model of asthma but dispensable for their differentiation and function. Mediators Inflamm. 2019:1656484. DOI: 10.1155/2019/1656484. PMID: 31178661. PMCID: PMC6507252. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85067541537&origin=inward.

36. Ghonim MA, Pyakurel K, Ibba SV, Wang J, Rodriguez P, Al-Khami AA, Lammi MR, Kim H, Zea AH, Davis C, Okpechi S, Wyczechowska D, Al-Ghareeb K, Mansy MS, Ochoa A, Naura AS, Boulares AH. 2015; PARP is activated in human asthma and its inhibition by olaparib blocks house dust mite-induced disease in mice. Clin Sci (Lond). 129:951–962. DOI: 10.1042/CS20150122. PMID: 26205779. PMCID: PMC4613510. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84952012202&origin=inward.

37. Holguin F, Cardet JC, Chung KF, Diver S, Ferreira DS, Fitzpatrick A, Gaga M, Kellermeyer L, Khurana S, Knight S, McDonald VM, Morgan RL, Ortega VE, Rigau D, Subbarao P, Tonia T, Adcock IM, Bleecker ER, Brightling C, Boulet LP, et al. 2020; Management of severe asthma: a European Respiratory Society/American Thoracic Society guideline. Eur Respir J. 55:1900588. DOI: 10.1183/13993003.00588-2019. PMID: 31558662. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85077155298&origin=inward.

38. Saglani S. 2017; Childhood severe asthma: New insights on remodelling and biomarkers. Paediatr Respir Rev. 24:11–13. DOI: 10.1016/j.prrv.2017.06.001. PMID: 28697969. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85022059264&origin=inward.

Fig. 1
PARP-1 inhibits the migration of 16HBE cells.
(A, B) Western blot and qRT-PCR were performed to evaluate the protein and mRNA expressions of PARP-1 in 16HBE cells transfected with NC, PARP-1 plasmid, or PARP-1 siRNA after 48 h. The experiments were replicated three times and presented as mean ± SD. *p < 0.05 vs. NC group. (C) Effects of PARP-1 on migration in 16HBE cells were detected by Transwell assay. Scale bars = 50 μm. The experiments were replicated three times and presented as mean ± SD. PARP-1, poly (ADP-ribose) polymerase-1; 16HBE, Human bronchial epithelial cell; NC, normal control; siRNA, small interfering RNA; siPARP-1, PARP-1 siRNA. *p < 0.05 vs. NC group, **p < 0.01 vs. NC group.
Fig. 2
MiR-21 suppresses PARP-1 expression and PARP-1 is a direct target of miR-21.
(A–C) The 16HBE cells were transfected with NC, miR-21 mimics, miR-21 inhibitor, miR-21 mimics + PARP-1 plasmid, or miR-21 inhibitor + PARP-1 siRNA. Expressions of miR-21 and PARP-1 mRNA were detected by qRT-PCR and the protein expression of PARP-1 was detected by Western blot 48 h post-transfection. The experiments were replicated three times and presented as mean ± SD. *p < 0.05 vs. NC group, **p < 0.01 vs. NC group, †p < 0.05 vs. miR21 or miR-21 inhibitor group, ††p < 0.01 vs. miR-21 inhibitor group. (D) The potential binding site between miR-21 and 3’UTR of PARP-1 mRNA is shown. Luciferase activities of 16HBE cells transfected with NC, miR-21 mimics, PARP-1 Wt, PARP-1 Mut, miR-21 mimics + PARP-1 Wt, or miR-21 mimics + PARP-1 Mut were measured by luciferase reporter assay. The experiments were replicated three times and presented as mean ± SD. **p < 0.01 vs. NC group. (E) Western blot was used to detect the binding capacity of biotin-labeled miR-21 mimics probes with PARP-1 protein. The experiments were replicated three times. miR-21, microRNA-21; PARP-1, poly (ADP-ribose) polymerase-1; 16HBE, Human bronchial epithelial cell; NC, normal control; siRNA, small interfering RNA; siPARP-1, PARP-1 siRNA; UTR, untranslated region; Wt, wild type; Mut, mutant.
Fig. 3
Effects of miR-21/PARP-1 on migration of 16HBE cells.
(A, B) The 16HBE cells were transfected with NC, miR-21 mimics, miR-21 inhibitor, miR-21 mimics + PARP-1 plasmid, or miR-21 inhibitor + PARP-1 siRNA. Transwell assay was performed to assess the effects of miR-21/PARP-1 on the migration of 16HBE cells. Scale bars = 50 μm. The experiments were replicated three times and presented as mean ± SD. ***p < 0.001 vs. NC group, †††p < 0.001 vs. miR21 inhibitor group, ††††p < 0.0001 vs. miR21 group. (C) Images of the cell morphology 72 h post transfection. Scale bars = 50 μm. miR-21, microRNA-21; PARP-1, poly (ADP-ribose) polymerase-1; 16HBE, Human bronchial epithelial cell; NC, normal control; siRNA, small interfering RNA; siPARP-1, PARP-1 siRNA.

Fig. 4
Effects of PARP-1 on expression of EMT-related proteins and EMT-regulating transcription factors.
(A, C) Western blot analysis evaluated the expression of Snail, ZEB1, N-cadherin, Vimentin and E-cadherin in 16HBE cells transfected with NC, PARP-1 plasmid, or PARP-1 siRNA after 48 h. The experiments were replicated three times and presented as mean ± SD. *p < 0.05 vs. NC group, **p < 0.01 vs. NC group. (B, D) The mRNA expressions of Snail, ZEB1, N-cadherin, Vimentin, and E-cadherin in 16HBE cells transfected with NC, PARP-1 plasmid, or PARP-1 siRNA after 48 h, as detected by qRT-PCR. The experiments were replicated three times and presented as mean ± SD. PARP-1, poly (ADP-ribose) polymerase-1; EMT, epithelial-mesenchymal transition; 16HBE, Human bronchial epithelial cell; NC, normal control; siRNA, small interfering RNA; siPARP-1, PARP-1 siRNA. *p < 0.05 vs. NC group, **p < 0.01 vs. NC group, ***p < 0.001 vs. NC group.
Fig. 5
Overexpression of PARP-1 alleviates the EMT induced by miR-21 mimics in 16HBE cells.
(A, C, E, G) Western blot analysis evaluated the protein expression of Snail, ZEB1, N-cadherin, Vimentin, and E-cadherin in 16HBE cells transfected with NC, miR-21 mimics, miR-21 inhibitor, miR-21 mimics + PARP-1 plasmid or miR-21 inhibitor + PARP-1 siRNA after 48 h. The experiments were replicated three times and presented as mean ± SD. *p < 0.05 vs. NC group, **p < 0.01 vs. NC group, †p < 0.05 vs. miR21 or miR21 inhibitor group, ††p < 0.01, vs. miR21 or miR21 inhibitor group. (B, D, F, H) The mRNA expression of Snail, ZEB1, E-cadherin, Vimentin, and E-cadherin in 16HBE cells transfected with NC, miR-21 mimics, miR-21 inhibitor, miR-21 mimics + PARP-1 plasmid, or miR-21 inhibitor + PARP-1 siRNA after 48 h were detected by qRT-PCR. The experiments were replicated three times and presented as mean ± SD. PARP-1, poly (ADP-ribose) polymerase-1; EMT, epithelial-mesenchymal transition; miR-21, microRNA-21; 16HBE, Human bronchial epithelial cell; NC, normal control; siRNA, small interfering RNA; siPARP-1, PARP-1 siRNA. *p < 0.05 vs. NC group, **p < 0.01 vs. NC group, ***p < 0.001 vs. NC group, ****p < 0.0001 vs. NC group, †p < 0.05 vs. miR21 inhibitor group, ††p < 0.01 vs. miR21 group, †††p < 0.001 vs. miR21 group.
Fig. 6
Effects of miR-21/PARP-1 on the PI3K/AKT signaling pathway.
(A–C) Western blot analysis evaluated the expression of PI3K/AKT-signaling-pathway-related molecular proteins in 16HBE cells transfected with NC, miR-21 mimics, miR-21 inhibitor, PARP-1 plasmid, PARP-1 siRNA, miR-21 mimics + PARP-1 plasmid, or miR-21 inhibitor + PARP-1 siRNA after 48 h. The experiments were replicated three times and presented as mean ± SD. PARP-1, poly (ADP-ribose) polymerase-1; 16HBE, Human bronchial epithelial cell; NC, normal control; miR-21, microRNA-21; siRNA, small interfering RNA; siPARP-1, PARP-1 siRNA. *p < 0.05 vs. NC group, **p < 0.01 vs. NC group, ***p < 0.001 vs. NC group, †p < 0.05 vs. miR21 or miR-21 inhibitor group, ††p < 0.01 vs. miR21 inhibitor group.
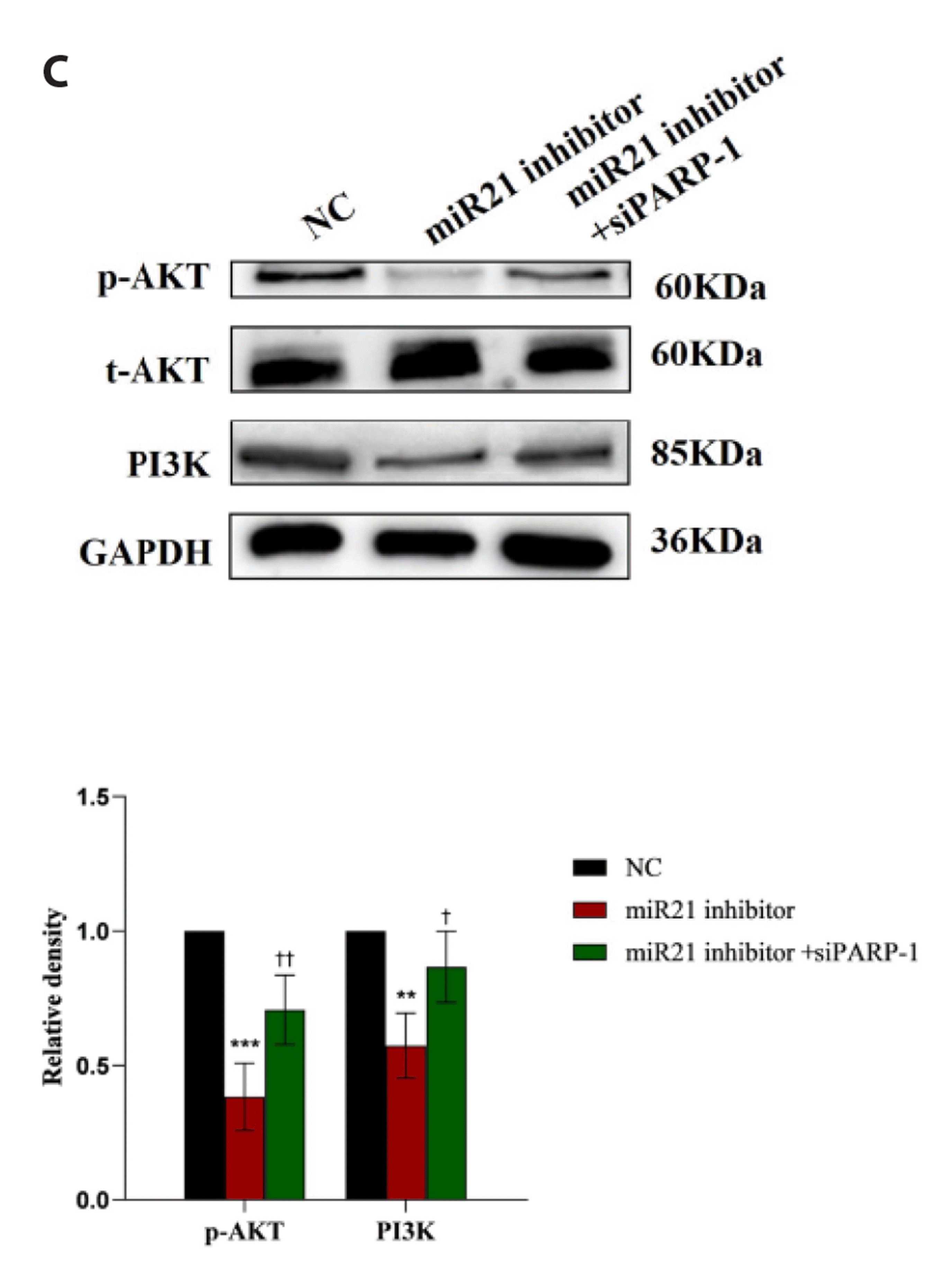
Fig. 7
PI3K inhibitor LY294002 inhibits the migration of 16HBE cells.
(A, B) The PI3K inhibitor LY294002 was used to study effects on the migration of 16HBE cells transfected with NC, miR-21 mimics, or PARP-1 siRNA. The experiments were replicated three times and presented as mean ± SD. 16HBE, Human bronchial epithelial cell; NC, normal control; miR-21, microRNA-21; PARP-1, poly (ADP-ribose) polymerase-1; siRNA, small interfering RNA; siPARP-1, PARP-1 siRNA; D, dimethylsulfoxide; LY, LY294002. Scale bars = 50 μm. *p < 0.05 vs. NC + D group, ***p < 0.001 vs. NC + D group, ††p < 0.01 vs. siPARP-1 + D group, †††p < 0.001 vs. miR21 + D group.

Fig. 8
MiR-21/PARP-1 regulates the EMT of 16HBE cells via activation of the PI3K/AKT signaling pathway.
(A, B) Western blot analysis evaluated the protein expressions of PI3K/AKT-signaling-pathway-related molecular proteins in 16HBE cells transfected with NC, miR-21 mimics, or PARP-1 siRNA after 48 h. The experiments were replicated three times and presented as mean ± SD. *p < 0.05 vs. NC + D group, **p < 0.001 vs. NC + D group, †p < 0.05 vs. miR21 + D or siPARP-1 + D group. (C–F) Effects of LY294002 on the EMT-related proteins in 16HBE cells transfected with NC, miR-21 mimics, or PARP-1 siRNA, as detected by Western blot and qRT-PCR after 48 h. The experiments were replicated three times and presented as mean ± SD. miR-21, microRNA-21; PARP-1, poly (ADP-ribose) polymerase-1; EMT, epithelial-mesenchymal transition; 16HBE, Human bronchial epithelial cell; NC, normal control; siRNA, small interfering RNA; siPARP-1, PARP-1 siRNA; D, dimethylsulfoxide; ns, not significant; LY, LY294002. *p < 0.05 vs. NC + D group, **p < 0.01 vs. NC + D group, ***p < 0.001 vs. NC + D group, ****p < 0.0001 vs. NC + D group, †p < 0.05 vs. miR21 + D or siPARP-1 + D group, ††p < 0.01 vs. miR21 + D or siPARP-1 + D group, †††p < 0.001 vs. miR21 + D or siPARP-1 + D group.
Fig. 9
Schematic diagram illustrating how miR-21/PARP-1 activates the PI3K/AKT signaling pathway to induce EMT and migration of 16HBE cells.
miR-21, microRNA-21; PARP-1, poly (ADP-ribose) polymerase-1; EMT, epithelial-mesenchymal transition; 16HBE, Human bronchial epithelial cell.
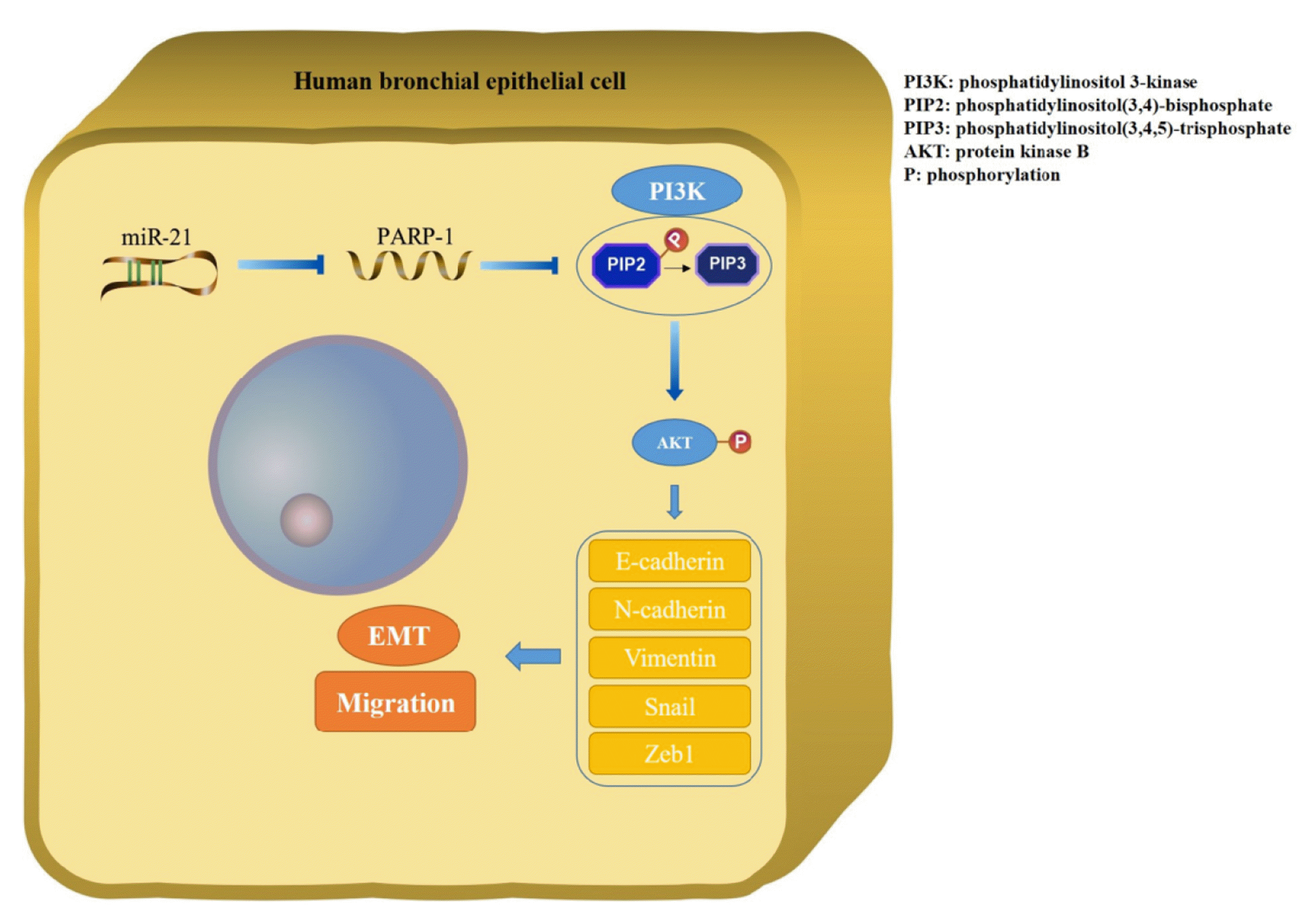
Table 1
Specific primer sequences for qRT-PCR




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download