Abstract
Notes
AUTHOR CONTRIBUTIONS
Lee DS and Lee J designed the study and wrote the manuscript. Lee DS and Roh EY collected the samples. Lee DS, Lee J, and Yun J reviewed the medical records of the patients. Kim S performed the cytogenetic analyses. Kim SM processed the data. Yun J, Jeong D, and Lee Y interpreted the data. Lee DS contributed to the revision of the manuscript. All authors approved the final manuscript to be published.
REFERENCES
Fig. 1
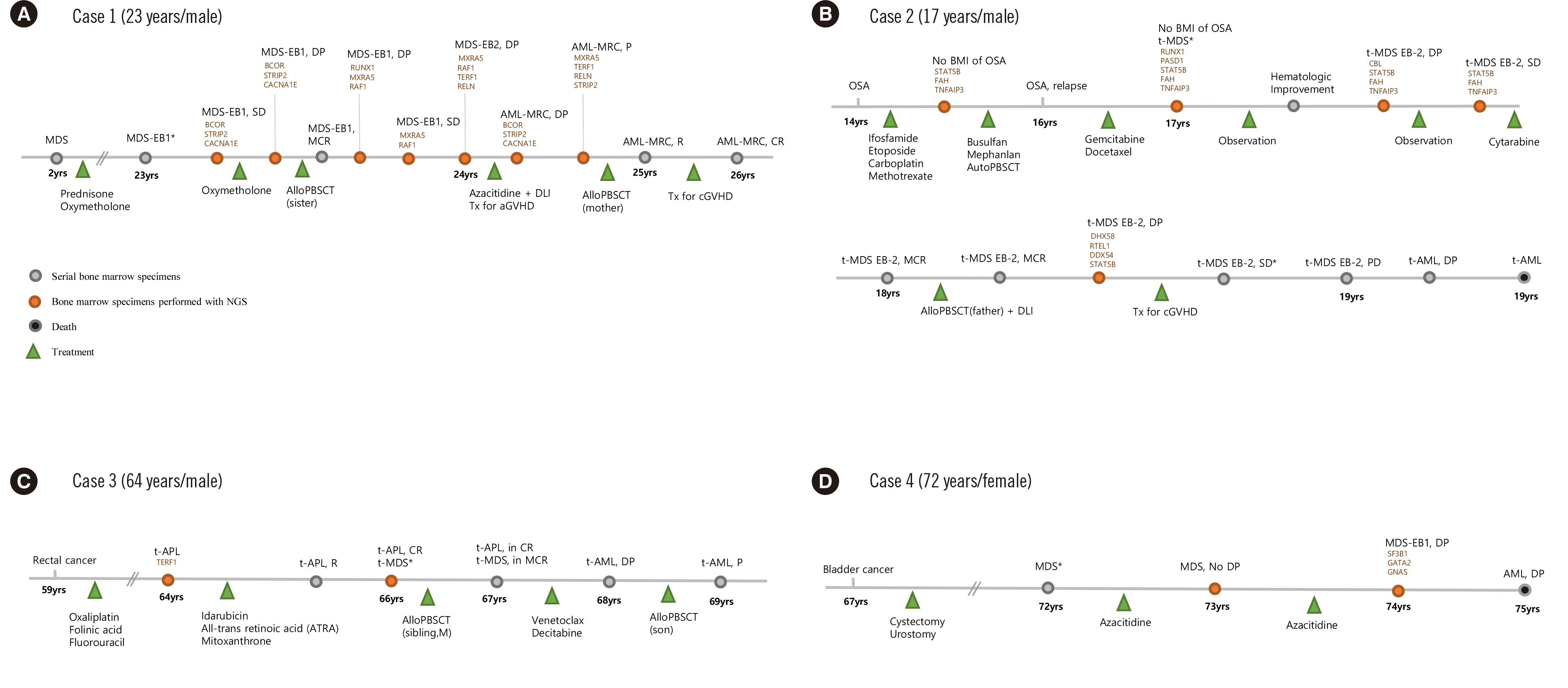
Table 1
| Characteristics | Case 1 | Case 2 | Case 3 | Case 4 |
|---|---|---|---|---|
| Diagnosis* | MDS-EB1 | t-MDS | t-MDS | MDS-U |
| Age† (yr)/sex | 23/male | 17/male | 66/male | 72/female |
| Underlying disease (age, yr) | MDS (2) | Osteosarcoma (16) | Rectal cancer (59) | Bladder cancer (67) |
| Chemotherapy or RT | None | Methotrexate, ifosfamide, etoposide, carboplatin, busulfan, melphalan | Oxaliplatin, folinic acid, fluorouracil | None |
| Survival‡ | 78 months (alive) | 31 months | 37 months (alive) | 36 months |
| CBC (Hb, WBC, PLT) | 60 g/L, 1,800 × 106/L, 60 × 109/L | 119 g/L, 2,980 × 106/L, 73 × 109/L | 117 g/L, 2,130 × 106/L, 47 × 109/L | 74 g/L, 900 × 106/L, 48 × 109/L |
| Blast count in BM§ | 9.0% | < 5% | < 5% | < 5% |
| Dysplasia | Dysgranulopoiesis | Dyserythropoiesis, dysmegakaryopoiesis | Dysmegakaryopoiesis | N/A |
| Chromosome (G-banding)ll | 46,XY,t(3;21)(q26.2;q22) | 45,XY,t(3;21)(q26.2;q22),–7 | 46,XY,t(3;21)(q26.2;q22) | 46,XX,t(3;21)(q26.2;q22) |
| MECOM FISH positivityll | Positive (52.7%) | Positive (46%) | Positive (50%) | N/A |
| Somatic variant genes (VAF, %) | RUNX1 (16.1) | RUNX1 (43.6) | SF3B1 (23.9) | |
| BCOR (62.1) | DHX58 (13.0) | TERF1 (17.3) | GATA2 (27.9) | |
| MXRA5 (48.9) | RTEL1 (44.1) | GNAS (22.6) | ||
| RAF1 (38.8) | DDX54 (57.3) | |||
| TERF1 (12.3) | CBL (57.7) | |||
| RELN (22.5) | PASD1 (14.5) | |||
| STRIP2 (49.5) | STAT5B (73.5) | |||
| CACNA1E (39.4) | FAH (50.0) | |||
| TNFAIP3 (46.4) |
*Initial hematologic diagnosis in the presence of a MECOM rearrangement; †Age at initial hematologic diagnosis with MECOM rearrangement; ‡Survival time from initial hematologic diagnosis to April 2021 for patients who are still alive; §Blast count observed on BM aspiration or BM section at initial diagnosis; llChromosome and MECOM FISH results at AML transformation.
Abbreviations: MDS, myelodysplastic syndrome; MDS-EB1, myelodysplastic syndrome with excess blasts 1; t-MDS, treatment-related myelodysplastic syndrome; MDS-U, myelodysplastic syndrome, unclassifiable; RT, radiotherapy; CBC, complete blood count; BM, bone marrow; N/A, not available due to poor quality; VAF, variant allele frequency; WBC, white blood cell; PLT, platelet.
Table 2
| Case | No. | Chr | Start | End | Ref | Variant | Gene | Type | Accession No. | Base change | AA change | SIFT† | Polyphen2† | CADD‡ | Tier [10] |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 21 | 36,164,610 | 36,164,610 | T | G | RUNX1* | Substitution | NM_001001890 | c.1184A >C | p.Glu395Ala | D | B | 23.5 | 2 |
| 2 | X | 39,921,998 | 39,921,998 | C | T | BCOR | Substitution | NM_001123383 | c.4071+1G >A | p? | . | . | 25.2 | 2 | |
| 3 | X | 3,235,214 | 3,235,214 | C | A | MXRA5 | Substitution | NM_015419 | c.6508G >T | p.Ala2170Ser | D | D | 25.8 | 3 | |
| 4 | 3 | 12,645,774 | 12,645,774 | T | C | RAF1 | Substitution | NM_001354695 | c.353A >G | p.Tyr118Cys | T | P | 14.16 | 3 | |
| 5 | 8 | 73,921,284 | 73,921,286 | GAG | - | TERF1 | Deletion | NM_003218 | c.186_188del | p.Glu62del | . | . | . | 3 | |
| 6 | 7 | 103,236,929 | 103,236,929 | C | G | RELN | Substitution | NM_005045 | c.3513G >C | p.Met1171Ile | T | P | 25.4 | 3 | |
| 7 | 7 | 129,094,012 | 129,094,012 | G | A | STRIP2 | Substitution | NM_001134336 | c.560G >A | p.Arg187Gln | D | D | 35 | 3 | |
| 8 | 1 | 181,546,987 | 181,546,987 | C | G | CACNA1E | Substitution | NM_000721 | c.598C >G | p.Leu200Val | D | D | 28.3 | 3 | |
| 2 | 9 | 21 | 36,164,610 | 36,164,610 | T | G | RUNX1* | Substitution | NM_001001890 | c.1184A >C | p.Glu395Ala | D | B | 23.5 | 2 |
| 10 | 17 | 40,255,767 | 40,255,767 | G | A | DHX58 | Substitution | NM_024119 | c.1613C >T | p.Ala538Val | T | P | 11.83 | 3 | |
| 11 | 20 | 62,325,796 | 62,325,796 | C | G | RTEL1 | Substitution | NM_001283010 | c.2395C >G | p.Leu799Val | D | D | 26.1 | 3 | |
| 12 | 12 | 113,603,723 | 113,603,723 | C | T | DDX54 | Substitution | NM_001111322 | c.1529G >A | p.Arg510His | T | P | 16.74 | 3 | |
| 13 | 11 | 119,077,232 | 119,077,232 | - | CACCAC | CBL | Duplication | NM_005188 | c.122_127dup | p.His41_His42dup | . | . | . | 3 | |
| 14 | X | 150,817,142 | 150,817,144 | GCT | - | PASD1 | Deletion | NM_173493 | c.706_708del | p.Ala236del | . | . | . | 3 | |
| 15 | 17 | 40,370,849 | 40,370,849 | C | T | STAT5B | Substitution | NM_012448 | c.881G >A | p.Arg294His | D | D | 34 | 3 | |
| 16 | 15 | 80,454,614 | 80,454,614 | C | T | FAH | Substitution | NM_000137 | c.391C >T | p.Arg131Trp | D | P | 24.2 | 3 | |
| 17 | 6 | 138,199,573 | 138,199,573 | G | C | TNFAIP3 | Substitution | NM_001270507 | c.991G >C | p.Asp331His | D | D | 24.7 | 3 | |
| 3 | 18 | 8 | 73,921,284 | 73,921,286 | GAG | - | TERF1 | Deletion | NM_003218 | c.186_188del | p.Glu62del | . | . | . | 3 |
| 4 | 19 | 2 | 198,266,834 | 198,266,834 | T | C | SF3B1 | Substitution | NM_012433 | c.2098A >G | p.Lys700Glu | D | D | 28 | 1 |
| 20 | 3 | 128,205,776 | 128,205,776 | G | C | GATA2 | Substitution | NM_001145661 | c.99C >G | p.Tyr33* | . | . | 37 | 1 | |
| 21 | 20 | 57,428,427 | 57,428,427 | C | G | GNAS | Substitution | NM_080425 | c.107C >G | p.Ala36Gly | D | B | 23.5 | 3 |
*RUNX1 (c.1184A>C, p.Glu395Ala) was detected in cases 1 and 2; †Protein-level prediction algorithms (SIFT, Polyphen2) are presented for the nonsynonymous variants. Tolerated and deleterious variants found in the SIFT prediction algorithm are annotated as T and D, respectively, and benign, possibly damaging, and probably damaging variants identified from Polyphen2 prediction are annotated as B, P, and D, respectively; ‡The prediction algorithm CADD can score human single nucleotide variants and short insertion/deletions. Variants with score above 10 to 20 indicate potential deleteriousness in CADD prediction.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download