Acute myeloid leukemia (AML) is a biologically complex hematopoietic disease characterized by maturation arrest and the expansion of immature cells within the bone marrow (BM). Chromosomal aberrations detected at diagnosis are regarded as the most important factors for predicting the clinical outcome. Conventional cytogenetic analysis at the time of diagnosis provides the most important clinical information in AML patients; however, 40%–50% of patients do not harbor clonal chromosomal aberrations [
1]. Most cases of cytogenetically normal AML (CN-AML) are currently categorized in the intermediate-risk group, which is quite heterogeneous [
2,
3]. Identifying recurrent molecular abnormalities has improved prognosis and has provided insight into the mechanisms of leukemogenesis in patients with CN-AML, leading to the discovery of novel therapeutic targets [
4]. However, many of the polymorphisms and expression patterns identified are still not well understood and require further analysis for determining their exact roles in CN-AML [
5].
Recently, several molecular markers that allow the prediction of outcome risk, even in patients with CN-AML, have been identified [
6]. The presence or absence of somatically acquired genetic alterations in CN-AML and other cytogenetic groups is currently an important consideration when performing risk assessment in these patients. Following the identification of these polymorphisms, patients with CN-AML are now recognized as a diverse group with distinct clinical outcomes. Both the WHO and European Leukemia Net classify recurrent molecular abnormalities as a complement to cytogenetics [
4,
7,
8]. The identification of new molecular markers is an important requirement for the implementation of precision medicine for CN-AML patients.
Prohibitins (PHBs) are ubiquitously expressed in mammalian cells and mediate transcriptional repression through nuclear hormone receptors by recruiting histone deacetylases. They are involved in regulating mitochondrial respiration and aging. PHBs are tumor suppressors and anti-proliferative cell-cycle regulators in normal cells. They are localized in the nucleus, cytosol, and mitochondria and have distinct functions depending on their cellular localization. PHBs are divided into types I and II based on their similarity to yeast PHB1 and PHB2, respectively [
9]. PHB alterations have been found in aging and cancer as well as in neurodegenerative, cardiac, and kidney diseases associated with mitochondrial impairments [
9]. PHB2 protein expression is aberrantly increased in leukemia cells in BM aspirates of patients with AML at initial diagnosis [
10]. This observation prompted us to investigate the clinical implications of PHB2 protein expression in CN-AML.
Between July 2004 and March 2012, 134 patients (males:females, 1:0.87) with CN-AML were enrolled. This study was approved by the Institutional Review Board of Chonnam National University Hwasun Hospital (Hwasun, Korea) (IRB No. 2019-001). Informed consent was obtained from all participating patients (
Supplemental Data Table S1). AML was diagnosed according to the 2016 WHO classification criteria; all patients enrolled received intensive remission induction therapy consisting of three days of idarubicin at 12 mg/m
2/day and seven days of cytarabine at 100 mg/m
2/day or N4-behenoyl-1-D-arabinofuranosylcytosine (300 mg/m
2/day for patients younger than 40 years, 200 mg/m
2/day for patients older than 40 years). Patients who achieved received three courses of high-dose cytarabine chemotherapy (3 g/m2 every 12 hours/day on days 1, 3, and 5) showed complete remission (CR). All patients had
de novo AML, and 45% of patients achieved CR. Thirty-eight percent of patients received stem cell transplantation (SCT). Using an automatic stainer (Benchmark GX; Ventana Medical Systems, Inc., Oro Valley, AZ) and polyclonal rabbit anti-human PHB2 antibodies (Novus Biologicals, Centennial, CO), PHB2 protein expression was assessed via immunohistochemical staining (IHCS) of paraffin-embedded BM sections at the initial diagnosis phase of CN-AML. PHB2-immunostained sections were scored using our published protocol [
1]. PHB2-IHCS was independently examined and scored by two experts. Results of equivocal cases were interpreted and determined by a third expert (
Fig. 1).
Survival analysis was performed in comparison with other prognostic factors, including age, NPM1 polymorphism, and FLT3 internal tandem duplication (FLT3-ITD). FLT3-ITD was analyzed using PCR and fragment analysis (LeukoStrat FLT3 Mutation kit; Invivoscribe, Inc., San Diego, CA, USA). NPM1 polymorphism was analyzed using a quantitative reverse-transcription PCR kit (Ipsogen NPM1 MutaQuant; Qiagen, Hilden, Germany). All statistical computations and graphical process were performed using R version 3.5.1. P≤0.05 (95% confidence interval) was considered significant. Overall survival (OS) was analyzed using Kaplan–Meier survival curve estimates and log-rank tests to compare differences in the distribution of survival between groups. Hazard ratios (HRs) were analyzed using Cox regression analysis examining age, sex, BM blast percentage, NPM1 polymorphism, FLT3-ITD, and the PHB2 protein expression status.
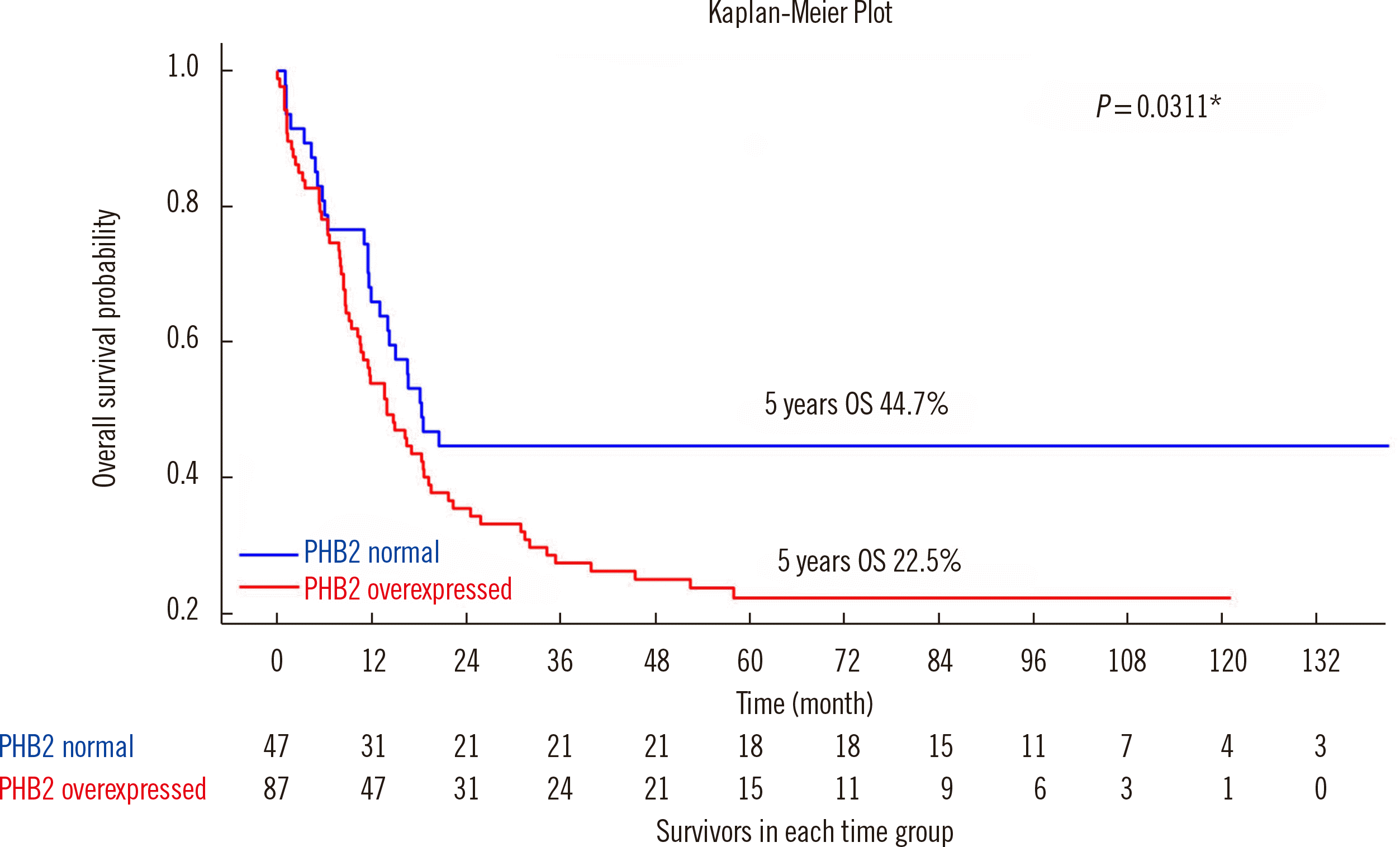
Table 1 shows the OS difference and HRs according to the different PHB2-IHCS score cutoffs. Among all scores (0–8), when we used 6 as a cutoff for PHB2 overexpression, the survival difference between the normal and overexpression groups was the highest (22.3%) and most significant, and the HR was significant (
P=0.033). Therefore, we selected 6 as the cutoff value. CN-AML patients with PHB2 protein overexpression showed inferior OS (22.5% vs. 44.7%;
P=0.031) and a high HR (1.64;
P=0.033) (
Table 1 and
Fig. 2). Among the well-known prognostic markers, none was associated with a distinct difference in OS between the patient groups (
Supplemental Data Figs. S1 and S2). Clinical characteristics, including age, sex, white blood cell (WBC) count in peripheral blood (PB), PB or BM blast percentage, CR achievement rate, or enforcement of SCT, were not significantly different according to the PHB2 levels estimated using IHCS at diagnosis (
Supplemental Data Table S1).
FLT3-ITD and
NPM1 polymorphism showed no clinical effect on OS. Aberrant
WT1 mRNA overexpression showed no significant clinical effect on OS. However, CN-AML patients with
BAALC mRNA overexpression had shorter OS (
Supplemental Data Fig. S1).
PHB is vital in tumorigenesis and tumor progression. However, its molecular role in cancer development has not been fully elucidated. Mitochondrial PHBs (mainly PHB2) are overexpressed in leukemia cells and promote cell survival under oxidative stress [
11]; however, the clinical implications thereof remained unclear. This result was consistent with ours that PHB2 is highly expressed in BM CN-AML cells. CN-AML patients with higher PHB2 expression in their AML cells showed worse OS than CN-AML patients having relatively lower PHB2 protein expression.
PHB protein levels are significantly higher in tumor cells isolated from patients with leukemia and lymphoma than in PB mononuclear cells from healthy donors. Concordant with PHB expression observed in tumor cell lines, PHB is overexpressed in primary lymphoid and myeloid tumor cells [
11]. These results suggest that PHBs are upregulated during tumorigenesis to maintain mitochondrial integrity; they may serve as novel biomarkers and molecular targets for therapeutic intervention of certain types of hematologic malignancies [
11].
The molecular mechanisms underlying effects of PHB2 in promoting cell migration have not been fully elucidated. Shen,
et al. [
12] reported that AKT (also known as protein kinase B) serine/threonine kinase 2 (AKT2) interacts with PHB2 in prostate cancer (PC) cell migration (PC3 and DU145). Combined overexpression of PHB2 and AKT2 inhibited the migration of both PC cell lines, suggesting that AKT2 overexpression abolished PHB2-induced migration [
12]. We previously investigated the molecular pathophysiological mechanisms of PHB in AML and found that PHB expression increased on inhibiting the mitogen-activated protein kinase pathway but decreased on activating the epidermal growth factor [
13]. Although cell proliferation signals downregulated the transcription of PHB, treatment with lithium chloride, an analog of Wnt signal, induced PHB expression in various cell types. Thus, we concluded that increased PHB expression in leukemia is regulated by Wnt signaling [
13].
In conclusion, PHB2 protein overexpression was associated with inferior OS in CN-AML patients. Assessing PHB2 protein expression in CN-AML using IHCS is a feasible approach to predicting disease activity and prognosis and may provide clinically useful information for the diagnosis, prognosis, and detection of minimal residual disease in CN-AML patients.





 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download