INTRODUCTION
MATERIALS AND METHODS
Patients
NGF-based MRD detection
Statistics
RESULTS
MRD status and patient characteristics
Table 1
| Total | MRD status* | |||
|---|---|---|---|---|
|
|
||||
| Negative | Positive | P | ||
| Patients (N) | 90 | 59 | 31 | |
| Sex, male | 52 (57.8%) | 34 (57.6%) | 18 (58.1%) | 0.968 |
| Age (yr) | 61 (55–67) | 61 (54–67) | 62 (57–67) | 0.425 |
| Myeloma type | ||||
| IgG | 44 (48.9%) | 27 (45.8%) | 17 (54.8%) | 0.887 |
| IgA | 16 (17.8%) | 11 (18.6%) | 5 (16.1%) | |
| IgD | 3 (3.3%) | 3 (5.1%) | 0 (0%) | |
| IgM | 1 (1.1%) | 1 (1.7%) | 0 (0%) | |
| Light chain only | 25 (27.8%) | 16 (27.1%) | 9 (29.0%) | |
| Non-secretary | 1 (1.1%) | 1 (1.7%) | 0 (0%) | |
| Light chain type | ||||
| Kappa | 54 (60.7%) | 34 (58.6%) | 20 (64.5%) | 0.587 |
| Lambda | 35 (39.3%) | 24 (41.4%) | 11 (35.5%) | |
| International staging system (N = 85) | ||||
| I | 25 (29.4%) | 16 (28.6%) | 9 (31.0%) | 0.957 |
| II | 29 (34.1%) | 19 (33.9%) | 10 (34.5%) | |
| III | 31 (36.5%) | 21 (37.5%) | 10 (34.5%) | |
| Cytogenetics (N = 84) | ||||
| Standard risk† | 64 (76.2%) | 45 (83.3%) | 19 (63.3%) | 0.039 |
| High risk‡ | 20 (23.8%) | 9 (16.7%) | 11 (36.7%) | |
| Treatment | ||||
| VTD | 59 (65.6%) | 38 (64.4%) | 21 (67.7%) | 0.847 |
| VMP | 12 (13.3%) | 9 (15.3%) | 3 (9.7%) | |
| Others | 19 (21.1%) | 12 (20.3%) | 7 (22.6%) | |
| ASCT | 70 (77.8%) | 46 (78%) | 24 (77.4%) | 1.000 |
| Response status at the time of MRD assessment (N = 89) | ||||
| sCR | 62 (69.7%) | 44 (71.0%) | 18 (29.0%) | 0.176 |
| CR | 12 (13.5%) | 7 (58.3%) | 5 (41.7%) | |
| VGPR | 15 (16.9%) | 7 (46.7%) | 8 (53.3%) | |
| Median follow-up duration after MRD assessment, months | 7 (3–12) | 6 (2–12) | 9 (3–14) | 0.993 |
| Progressive disease after MRD assessment | 15 (16.7%) | 5 (8.5%) | 10 (32.3%) | 0.004 |
*MRD status was based on the results of the last MRD assessment; †Other than high-risk cytogenetics; ‡del(17p), t(4;14)(p16;q32), and/or t(14;16)(q32;q23).
Abbreviations: MM, multiple myeloma; MRD, minimal residual disease; VTD, bortezomib-thalidomide-dexamethasone; VMP, bortezomib-melphalan-prednisone; ASCT, autologous stem cell transplantation; sCR, stringent complete remission; CR, complete remission; VGPR, very good partial response; IQR, interquartile range.
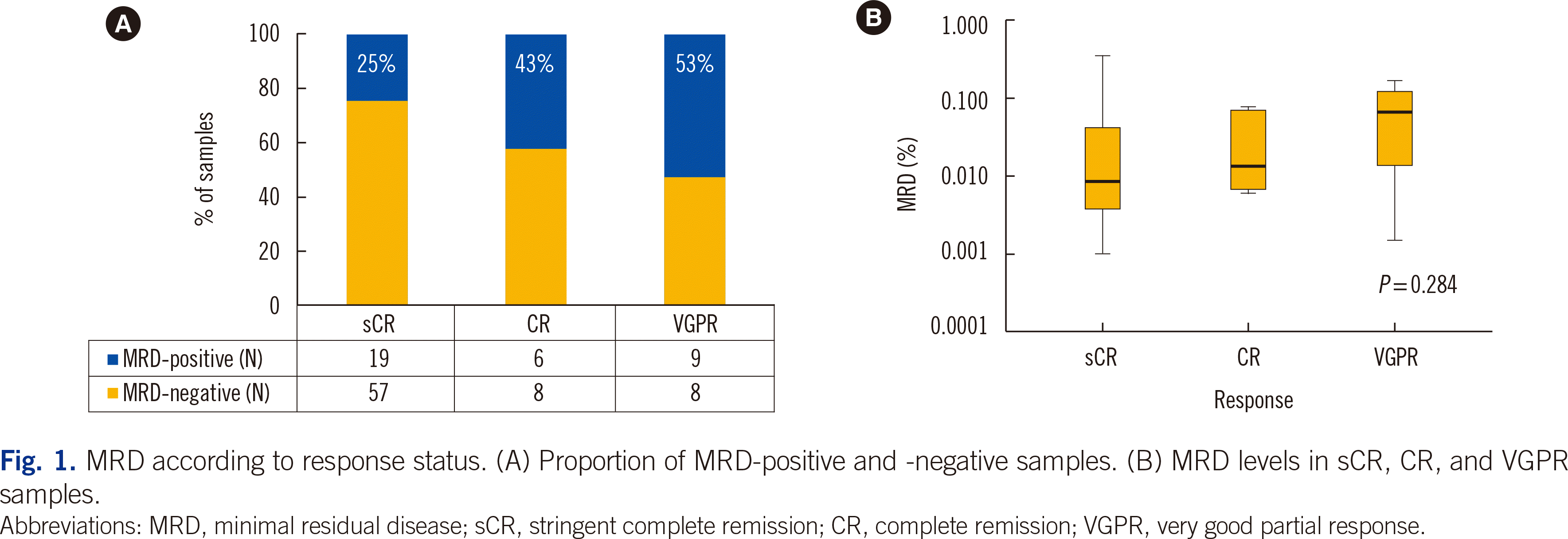
MRD status according to clinical response
Survival analysis according to clinical response and MRD
Fig. 2
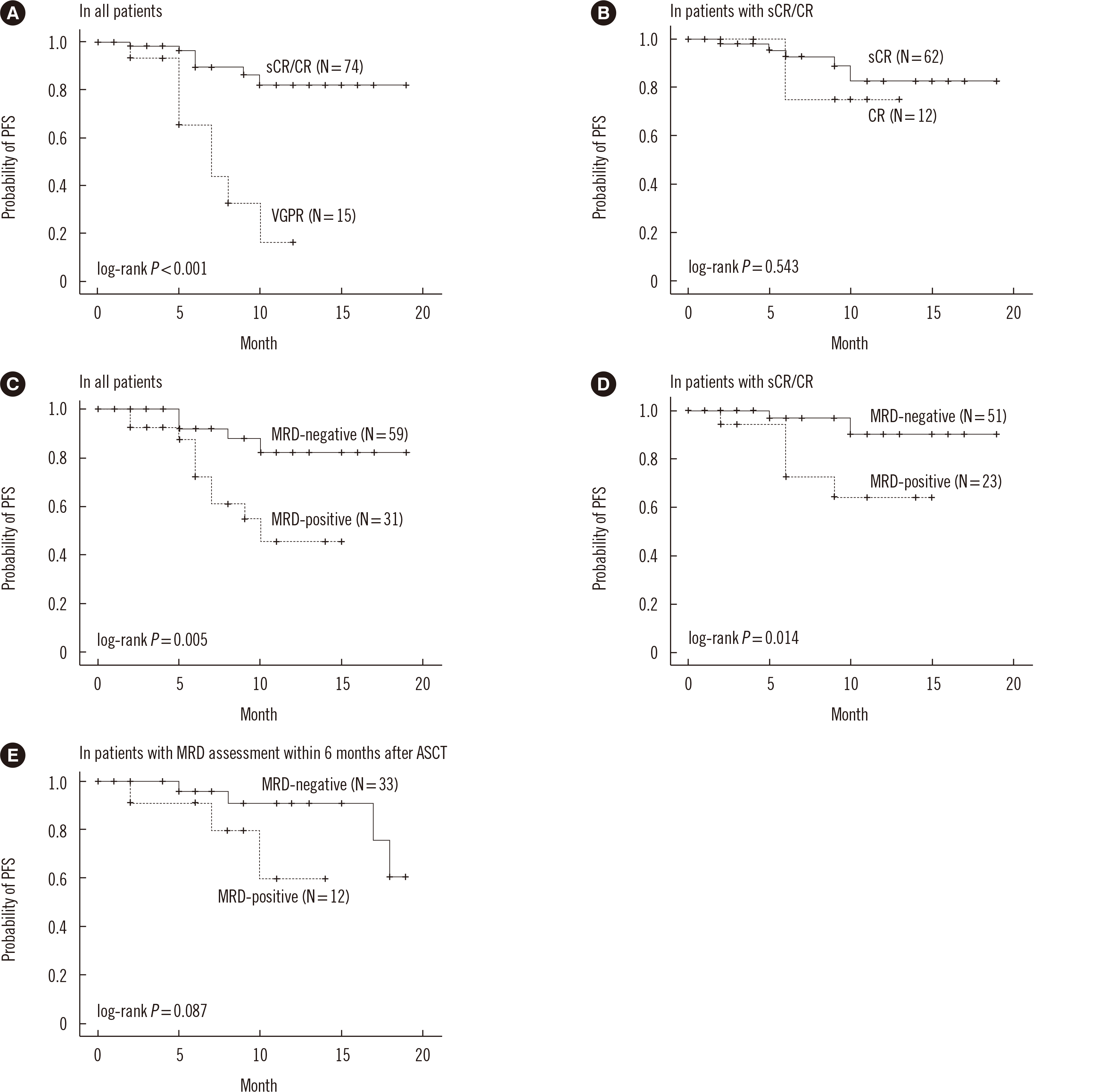
Table 2
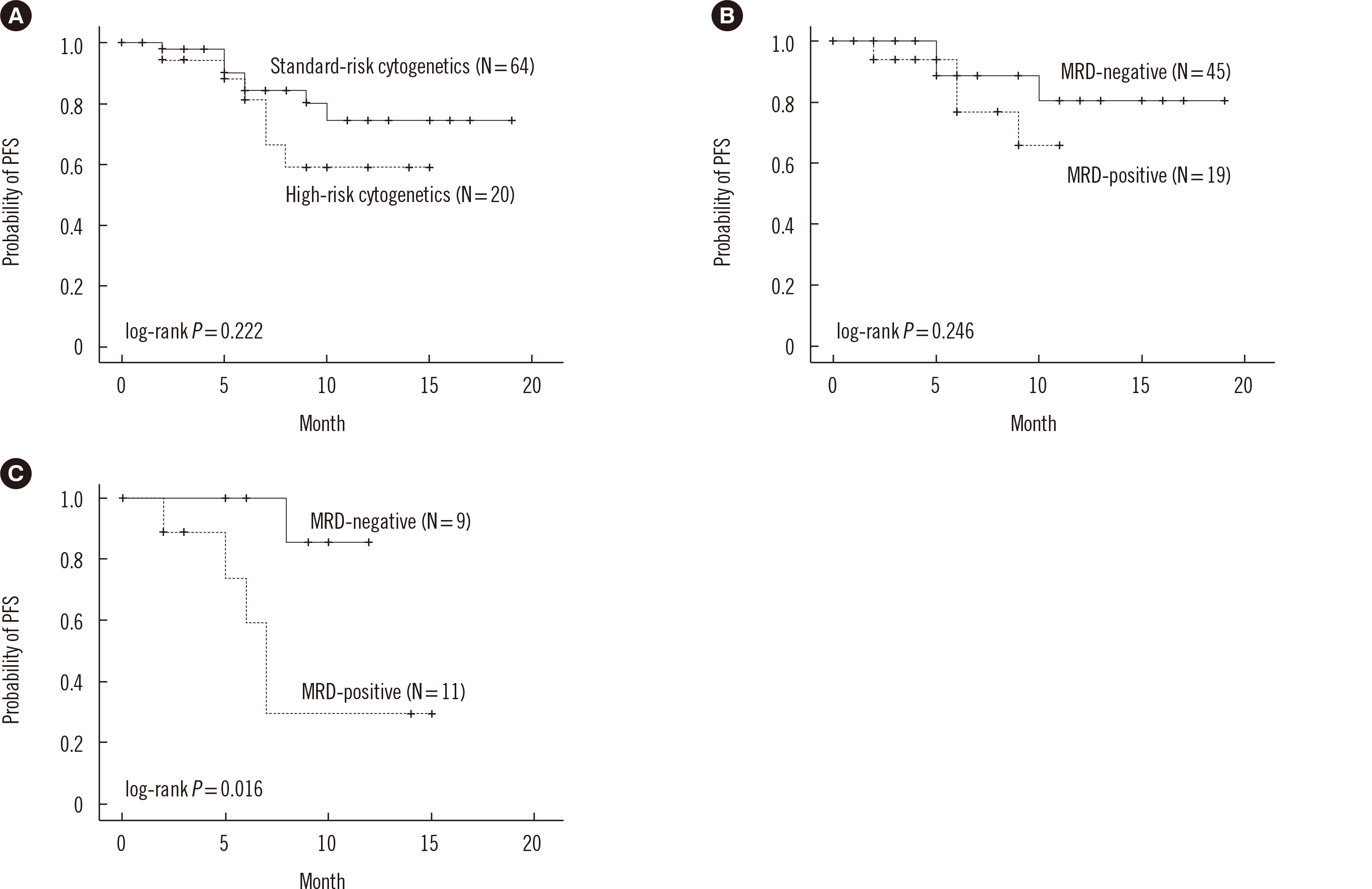
Survival analysis according to cytogenetic risk and MRD
Fig. 3
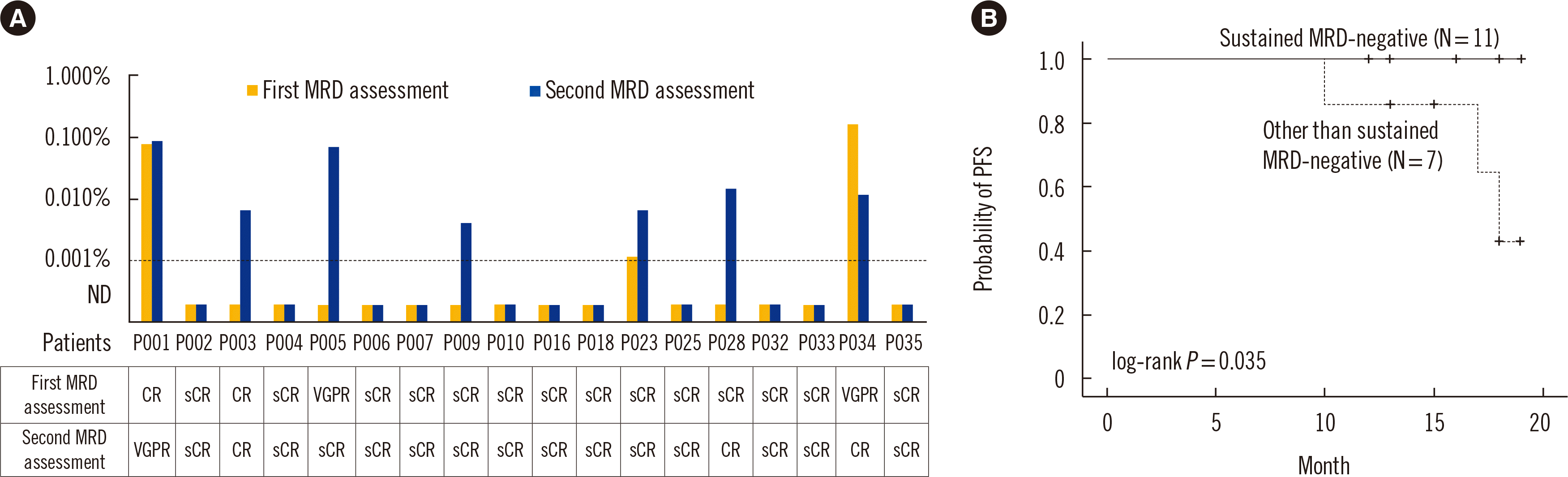
Patient characteristics and survival according to sustained MRD status
Fig. 4




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download