INTRODUCTION
METHODS
Clinical specimens
Cell culture and transfection
Reverse transcription quantitative polymerase chain reaction
Table 2
Western blot analysis
RNA pull down assay
Cell counting kit-8 assay
Wound healing assay
Statistical analyses
RESULTS
Expression of 10 mRNAs in plaques and adjacent intimal tissues from AS patients
 | Fig. 1Expression of 10 mRNAs in plaques and adjacent intimal tissues from atherosclerosis (AS) patients.(A) Expression levels of ten mRNAs (UBE2G2, SNORD45C, SNORD45A, SNORD45B, RABGGTB, SLC16A3, POLR2C, PNO1, CEMP1, AMDHD2) in plaques and adjacent intima from coronary AS patients were measured using RT-qPCR with pared t-test, as presented by the scatter diagram. (B) mRNAs (UBE2G2, SNORD45C, SNORD45A, SLC16A3, POLR2C, PNO1, CEMP1, AMDHD2) with significant difference were listed in the bar graph. *p < 0.05, **p < 0.01, NS indicates no significance.
|
Table 3
Expression of 10 mRNAs in serum from AS patients and healthy controls
 | Fig. 2Expression of 10 mRNAs in serum from atherosclerosis (AS) patients and healthy controls.(A) Expression levels of ten mRNAs (UBE2G2, SNORD45C, SNORD45A, SNORD45B, RABGGTB, SLC16A3, POLR2C, PNO1, CEMP1, AMDHD2) in the serum of coronary AS patients and healthy patients were measured using RT-qPCR with unpaired t-test. (B) mRNAs (UBE2G2, SNORD45C, SNORD45A, SLC16A3, POLR2C, PNO1, CEMP1, AMDHD2) with significant difference were listed. *p < 0.05, **p < 0.01, ***p < 0.001, NS indicates no significance.
|
Potential upstream miRNAs for the abnormally expressed mRNAs
 | Fig. 3Potential upstream miRNAs for the abnormally expressed mRNAs.(A) TargetScan predicted the potential upstream miRNAs of UBE2G2, SLC16A3, POLR2C, PNO1, and AMDHD2. (B) Expression levels of 11 miRNAs in plaques and adjacent intima from coronary AS patients were examined using RT-qPCR with pared t-test. (C) Expression levels of 11 miRNAs in the serum of coronary atherosclerosis (AS) patients and healthy patients were examined using RT-qPCR with unpaired t-test. *p < 0.05, **p < 0.01.
|
MiR-491-5p negatively regulates UBE2G2, SLC16A3, PNO1 expression in VSMCs
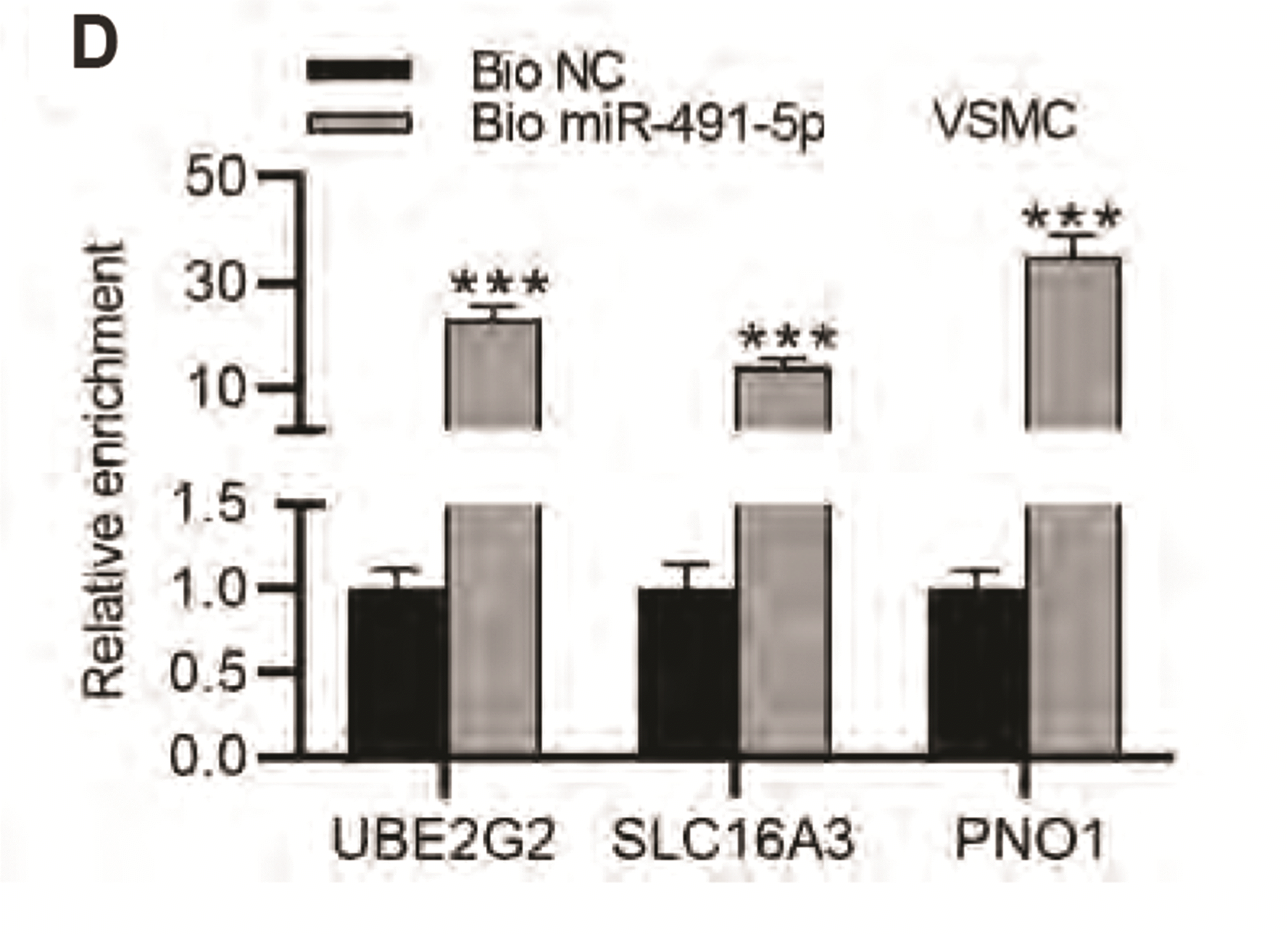 | Fig. 4MiR-491-5p negatively regulates UBE2G2, SLC16A3, PNO1 expression in vascular smooth muscle cells (VSMCs).(A) Overexpression efficiency of miR-491-5p in VSMCs was evaluated using RT-qPCR. (B) Expression levels of UBE2G2, SLC16A3, POLR2C, PNO1, and AMDHD2 in VSMCs transfected with miR-491-5p mimics and NC mimics were examined by RT-qPCR. (C) Protein levels of UBE2G2, SLC16A3, POLR2C, PNO1, and AMDHD2 in VSMCs transfected with miR-491-5p mimics and NC mimics were examined by Western blot. (D) RNA pull down assay was for detecting whether UBE2G2, SLC16A3, and PNO1 bind with miR-491-5p. *p < 0.05, **p < 0.01, ***p < 0.001.
|
MiR-491-5p inhibits viability, migration, and increased expression of contractile markers in VSMCs
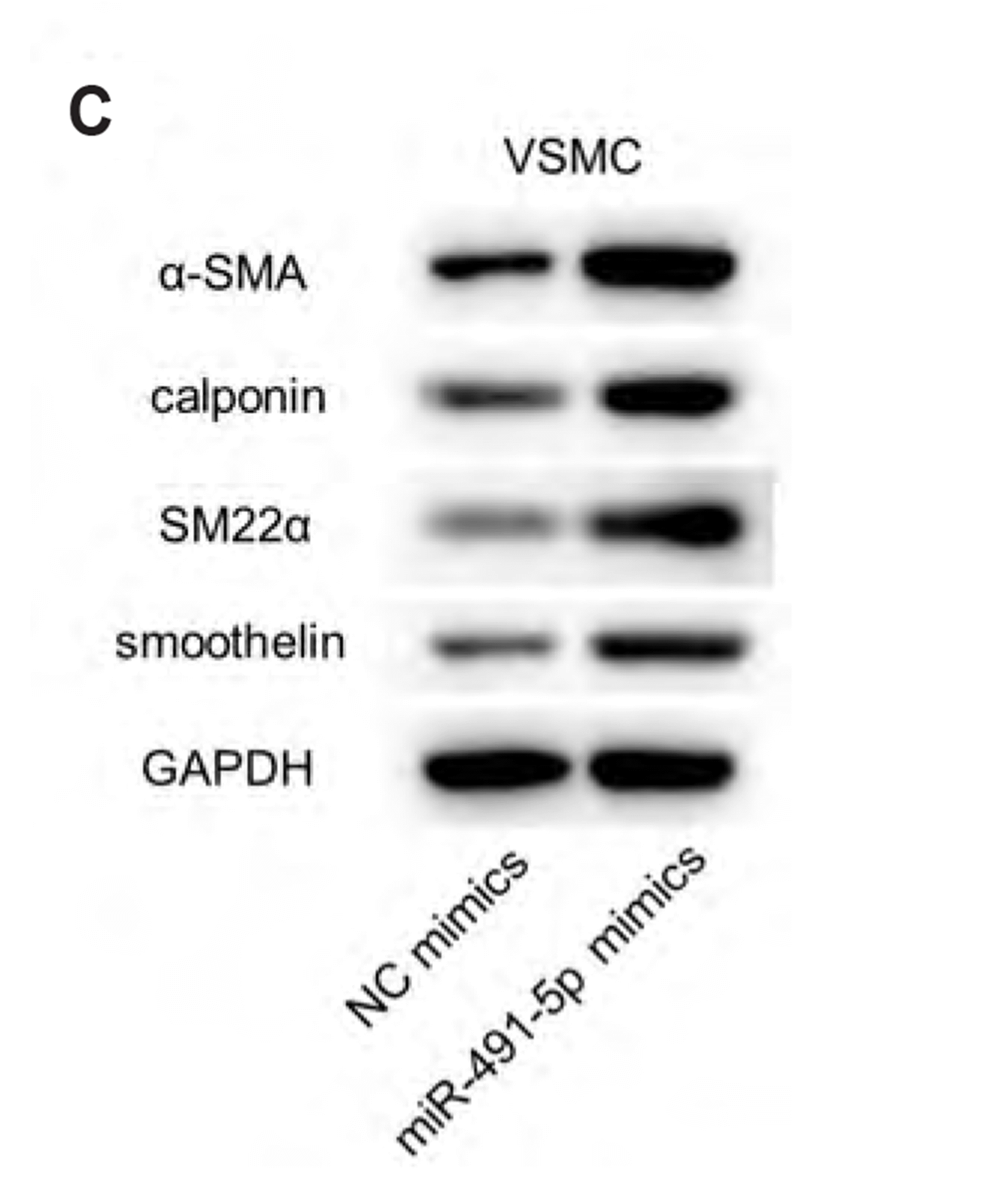 | Fig. 5MiR-491-5p inhibited viability, migration, and increased expression of contractile markers in vascular smooth muscle cells (VSMCs).(A) Cell counting kit 8 (CCK-8) revealed the viability in VSMCs with transfection of miR-491-5p mimics and NC mimics. (B) Wound healing was performed to detect the migration capacity of VSMCs with transfection of miR-491-5p mimics and NC mimics. (C) The protein levels of contractile markers (α-SMA, calponin, SM22α, and smoothelin) in VSMCs with transfection of miR-491-5p mimics and NC mimics were measured by Western blot. *p < 0.05, **p < 0.01.
|
MiR-491-5p antagomir promotes viability, migration, and increased expression of contractile markers in VSMCs
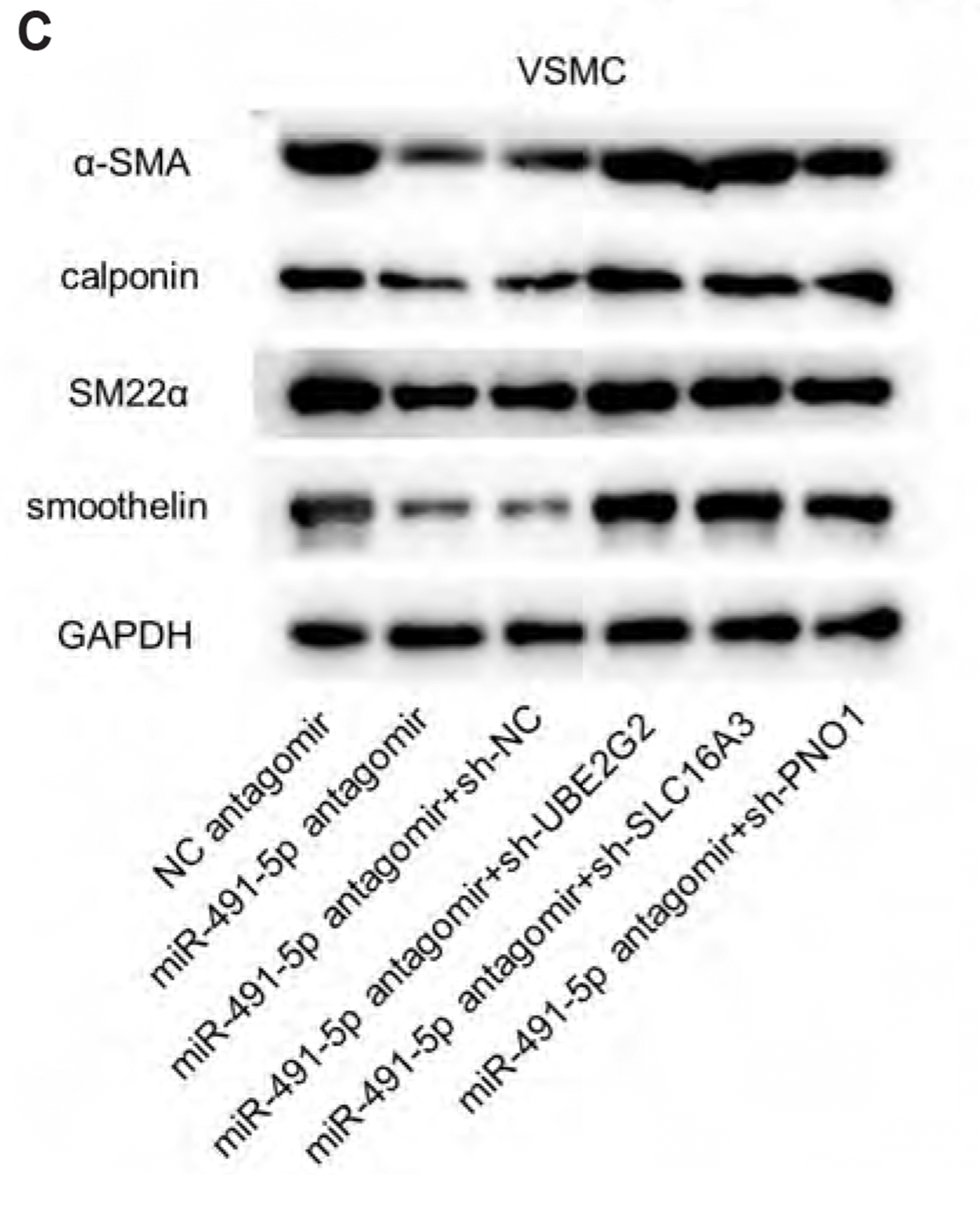 | Fig. 6MiR-491-5p antagomir promotes viability, migration, and increased expression of contractile markers in vascular smooth muscle cells (VSMCs).(A) CCK-8 revealed the viability in VSMCs with transfection of NC antagomir, miR-491-5p antagomir, miR-491-5p antagomir+sh-NC, miR-491-5p antagomir+sh-UBE2G2, miR-491-5p antagomir+sh-SLC16A3, and miR-491-5p antagomir+sh PNO1. (B) Wound healing was performed to detect the migration capacity of VSMCs with transfection of NC antagomir, miR-491-5p antagomir, miR-491-5p antagomir+sh-NC, miR-491-5p antagomir+sh-UBE2G2, miR-491-5p antagomir+sh- SLC16A3, and miR-491-5p antagomir+sh PNO1. (C) The protein levels of contractile markers (α-SMA, calponin, SM22α, and smoothelin) in VSMCs with transfection of NC antagomir, miR-491-5p antagomir, miR-491-5p antagomir+sh-NC, miR-491-5p antagomir+sh-UBE2G2, miR-491-5p antagomir+sh- SLC16A3, and miR-491-5p antagomir+sh PNO1. *p < 0.05, **p < 0.01.
|




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download