1. Fox JG, Wang TC. Inflammation, atrophy, and gastric cancer. J Clin Invest. 2007; 117:60–69.
2. Lim JH, Kim N, Lee HS, et al. Correlation between endoscopic and histological diagnoses of gastric intestinal metaplasia. Gut Liver. 2013; 7:41–50.
3. Panteris V, Nikolopoulou S, Lountou A, et al. Diagnostic capabilities of high-definition white light endoscopy for the diagnosis of gastric intestinal metaplasia and correlation with histologic and clinical data. Eur J Gastroenterol Hepatol. 2014; 26:594–601.
4. Dixon MF, Genta RM, Yardley JH, et al. Classification and grading of gastritis. The updated Sydney System. International Workshop on the Histopathology of Gastritis, Houston 1994. Am J Surg Pathol. 1996; 20:1161–1181.
5. Ang TL, Pittayanon R, Lau JY, et al. A multicenter randomized comparison between high-definition white light endoscopy and narrow band imaging for detection of gastric lesions. Eur J Gastroenterol Hepatol. 2015; 27:1473–1478.
6. Wu C, Namasivayam V, Li JW, et al. A prospective randomized tandem gastroscopy pilot study of linked color imaging versus white light imaging for detection of upper gastrointestinal lesions. J Gastroenterol Hepatol. 2021; 36:2562–2567.
7. Savarino E, Corbo M, Dulbecco P, et al. Narrow-band imaging with magnifying endoscopy is accurate for detecting gastric intestinal metaplasia. World J Gastroenterol. 2013; 19:2668–2675.
8. Pittayanon R, Rerknimitr R, Wisedopas N, et al. The learning curve of gastric intestinal metaplasia interpretation on the images obtained by probe-based confocal laser endomicroscopy. Diagn Ther Endosc. 2012; 2012:278045.
9. Sun M, Zhang G, Dang H, et al. Accurate gastric cancer segmentation in digital pathology images using deformable convolution and multi-scale embedding networks. IEEE Access. 2019; 7:75530–75541.
10. Li Y, Xie X, Liu S, et al. GT-Net: a deep learning network for gastric tumor diagnosis. In : In: 2018 IEEE 30th International Conference on Tools with Artificial Intelligence (ICTAI); 2018 Nov 5–7; Volos, Greece. p. 20–24.
11. Chen LC, Zhu Y, Papandreou G, et al. Encoder-decoder with atrous separable convolution for semantic image segmentation. In : In: Computer Vision (ECCV 2018); p. 833–851.
12. Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation. In : In: Medical Image Computing and Computer-Assisted Intervention (MICCAI 2015); p. 234–241.
13. Read P, Meyer MP. Restoration of motion picture film. Oxford: Butterworth-Heinemann;2000.
14. Rodriguez-Diaz E, Baffy G, Lo WK, et al. Real-time artificial intelligence-based histologic classification of colorectal polyps with augmented visualization. Gastrointest Endosc. 2021; 93:662–670.
15. Wang C, Li Y, Yao J, et al. Localizing and identifying intestinal metaplasia based on deep learning in oesophagoscope. In : In: 2019 8th International Symposium on Next Generation Electronics (ISNE); 2019 Oct 9–10; Zhengzhou, China. p. 1–4.
16. Sun X, Zhang P, Wang D, et al. Colorectal polyp segmentation by U-Net with dilation convolution. In : In: 2019 18th IEEE International Conference on Machine Learning and Applications (ICMLA); 2019 Dec 16–19; Boca Raton, FL. p. 851–858.
17. Yosinski J, Clune J, Bengio Y, et al. How transferable are features in deep neural networks? In : In: NIPS 14: Proceedings of the 27th International Conference on Neural Information Processing Systems; 2014 Dec 8–13; Montreal, Canada. p. 3320–3328.
18. Russell BC, Torralba A, Murphy KP, et al. LabelMe: a database and web-based tool for image annotation. Int J Comput Vis. 2008; 77:157–173.
19. Bernal J, Sanchez FJ, Fernández-Esparrach G, et al. WM-DOVA maps for accurate polyp highlighting in colonoscopy: validation vs. saliency maps from physicians. Comput Med Imaging Graph. 2015; 43:99–111.
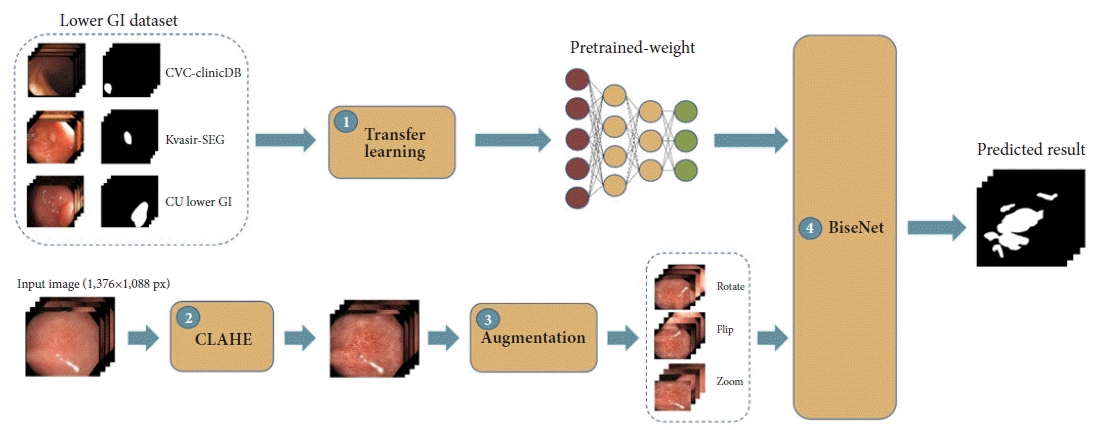
20. Jha D, Smedsrud PH, Riegler MA, et al. Kvasir-SEG: a segmented polyp dataset. In : In: MultiMedia Modeling: 26th International Conference, MMM 2020; 2020 Jan 5–8; Daejeon, Korea. p. 451–462.
21. Zuiderveld K. Contrast limited adaptive histogram equalization. In : Heckbert PS, editor. Graphics gems IV. San Diego (CA): Academic Press;1994. p. 474–485.
22. Yu C, Wang J, Peng C, et al. BiSeNet: bilateral segmentation network for real-time semantic segmentation. In : In: Computer Vision-ECCV 2018; p. 334–349.
23. Lecun Y, Bottou L, Bengio Y, et al. Gradient-based learning applied to document recognition. Proc IEEE. 1998; 86:2278–2324.
24. Goncalves W, Dos Santos M, Lobato F, et al. Deep learning in gastric tissue diseases: a systematic review. BMJ Open Gastroenterol. 2020; 7:e000371.
25. Mori Y, Neumann H, Misawa M, et al. Artificial intelligence in colonoscopy: now on the market. What’s next? J Gastroenterol Hepatol. 2021; 36:7–11.
26. Hirasawa T, Aoyama K, Tanimoto T, et al. Application of artificial intelligence using a convolutional neural network for detecting gastric cancer in endoscopic images. Gastric Cancer. 2018; 21:653–660.
27. Li L, Chen Y, Shen Z, et al. Convolutional neural network for the diagnosis of early gastric cancer based on magnifying narrow band imaging. Gastric Cancer. 2020; 23:126–132.
28. Zhang L, Zhang Y, Wang L, et al. Diagnosis of gastric lesions through a deep convolutional neural network. Dig Endosc. 2021; 33:788–796.
29. Suzuki H, Yoshitaka T, Yoshio T, et al. Artificial intelligence for cancer detection of the upper gastrointestinal tract. Dig Endosc. 2021; 33:254–262.
30. Xu M, Zhou W, Wu L, et al. Artificial intelligence in the diagnosis of gastric precancerous conditions by image-enhanced endoscopy: a multicenter, diagnostic study (with video). Gastrointest Endosc. 2021; 94:540–548.
31. He K, Zhang X, Ren S, et al. Deep residual learning for image recognition. In : In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2016 Jun 27–30; Las Vegas, NV. p. 770–778.
32. Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. In : In: 2015 International Conference on Learning Representations (ICLR); 2015 May 7–9; San Diego, CA.
33. Huang G, Liu Z, van der Maaten L, et al. Densely connected convolutional networks. In : In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2017 Jul 21–26; Honolulu, HI. p. 2261–2269.
34. Tan M, Le QV. EfficientNet: rethinking model scaling for convolutional neural networks. In : In: 36th International Conference on Machine Learning (ICML 2019); 2019 Jun 9–15; Long Beach, CA. p. 6105–6114.
35. ASGE Technology Committee, Abu Dayyeh BK, Thosani N, et al. ASGE Technology Committee systematic review and meta-analysis assessing the ASGE PIVI thresholds for adopting real-time endoscopic assessment of the histology of diminutive colorectal polyps. Gastrointest Endosc. 2015; 81:502. e1–502.e16.
36. Banks M, Graham D, Jansen M, et al. British Society of Gastroenterology guidelines on the diagnosis and management of patients at risk of gastric adenocarcinoma. Gut. 2019; 68:1545–1575.




 PDF
PDF Citation
Citation Print
Print




 XML Download
XML Download