1. WHO Classification of Tumours Editorial Board Central nervous system tumours. 5th ed. Lyon: International Agency for Research on Cancer;2021.
2. Broniscer A, Gajjar A. Supratentorial high-grade astrocytoma and diffuse brainstem glioma: two challenges for the pediatric oncologist. Oncologist. 2004; 9:197–206.

3. Hadjipanayis CG, Van Meir EG. Brain cancer propagating cells: biology, genetics and targeted therapies. Trends Mol Med. 2009; 15:519–30.

4. Paugh BS, Qu C, Jones C, et al. Integrated molecular genetic profiling of pediatric high-grade gliomas reveals key differences with the adult disease. J Clin Oncol. 2010; 28:3061–8.

5. Ohgaki H. Epidemiology of brain tumors. Methods Mol Biol. 2009; 472:323–42.

6. Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005; 352:987–96.

7. Richter A, Alexdottir MS, Magnus SH, et al. EGFL7 mediates BMP9-induced sprouting angiogenesis of endothelial cells derived from human embryonic stem cells. Stem Cell Reports. 2019; 12:1250–9.

8. Parker LH, Schmidt M, Jin SW, et al. The endothelial-cell-derived secreted factor Egfl7 regulates vascular tube formation. Nature. 2004; 428:754–8.

9. Fitch MJ, Campagnolo L, Kuhnert F, Stuhlmann H. Egfl7, a novel epidermal growth factor-domain gene expressed in endothelial cells. Dev Dyn. 2004; 230:316–24.

10. Hansen TF, Andersen RF, Olsen DA, Sorensen FB, Jakobsen A. Prognostic importance of circulating epidermal growth factor-like domain 7 in patients with metastatic colorectal cancer treated with chemotherapy and bevacizumab. Sci Rep. 2017; 7:2388.

11. Shen X, Han Y, Xue X, et al. Epidermal growth factor-like domain 7 promotes cell invasion and angiogenesis in pancreatic carcinoma. Biomed Pharmacother. 2016; 77:167–75.

12. Huang CH, Li XJ, Zhou YZ, Luo Y, Li C, Yuan XR. Expression and clinical significance of EGFL7 in malignant glioma. J Cancer Res Clin Oncol. 2010; 136:1737–43.

13. Wang FY, Kang CS, Wang-Gou SY, Huang CH, Feng CY, Li XJ. EGFL7 is an intercellular EGFR signal messenger that plays an oncogenic role in glioma. Cancer Lett. 2017; 384:9–18.

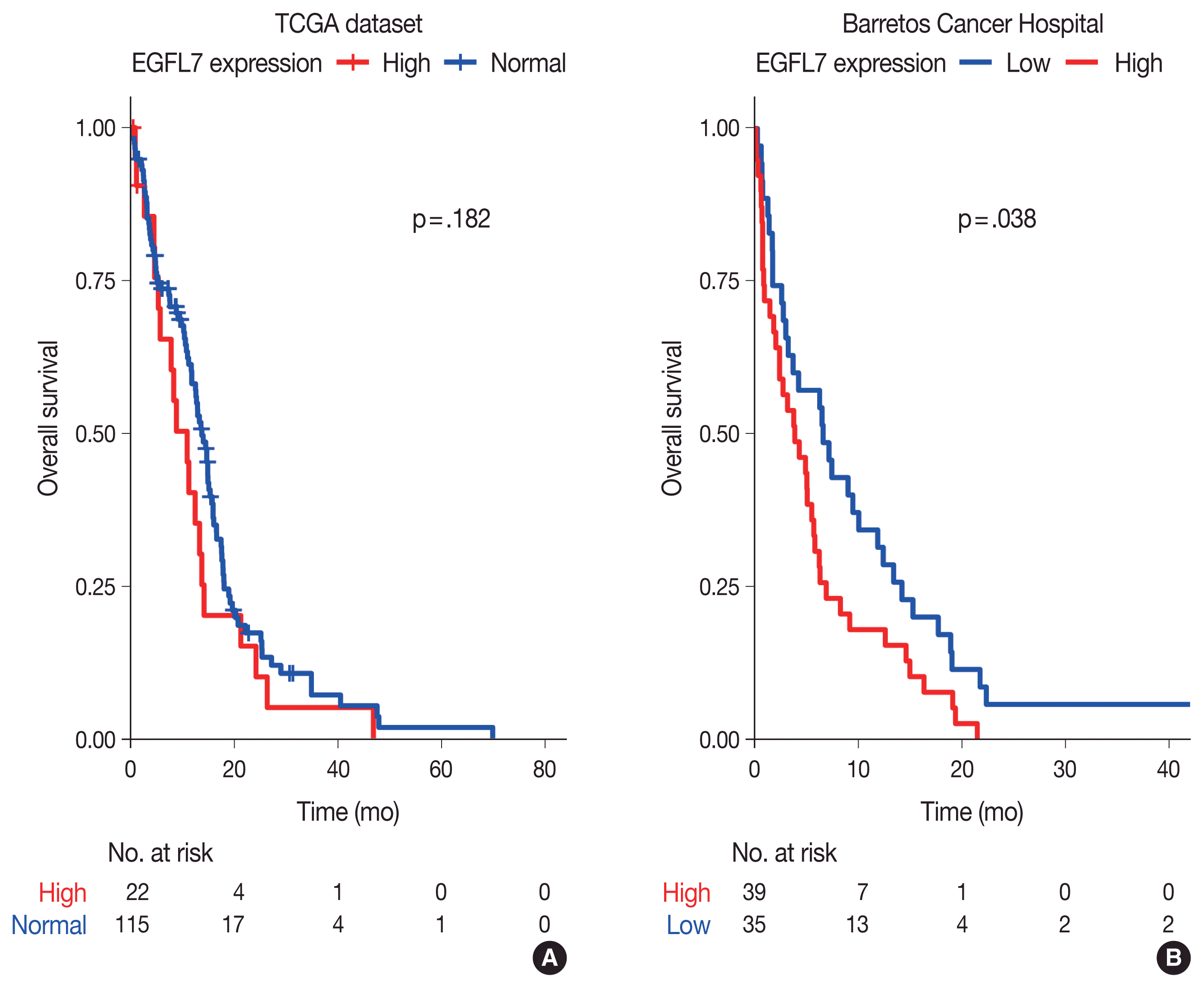
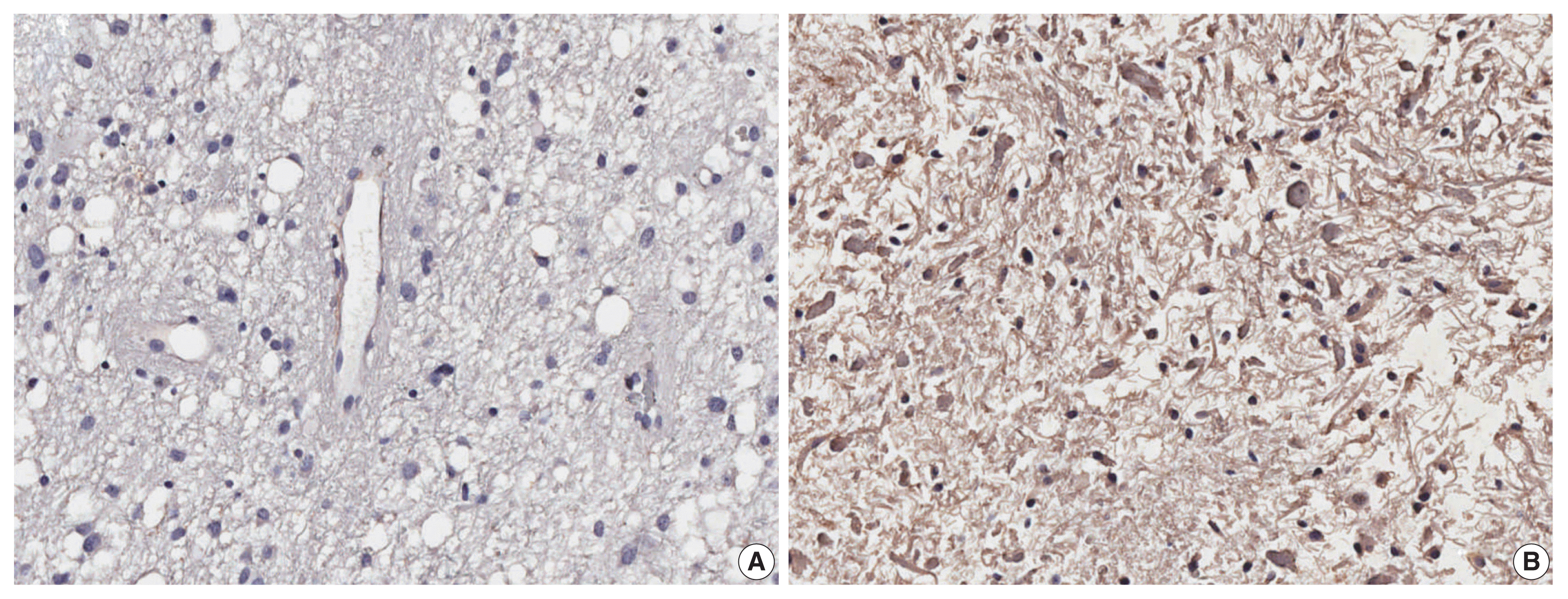
14. Brunhara BB, Becker AP, Neder L, et al. Evaluation of the prognostic potential of EGFL7 in pilocytic astrocytomas. Neuropathology. 2021; 41:21–8.

15. Fan C, Yang LY, Wu F, et al. The expression of Egfl7 in human normal tissues and epithelial tumors. Int J Biol Markers. 2013; 28:71–83.

16. Bidinotto LT, Torrieri R, Mackay A, et al. Copy number profiling of Brazilian astrocytomas. G3 (Bethesda). 2016; 6:1867–78.

17. Wan YW, Allen GI, Liu Z. TCGA2STAT: simple TCGA data access for integrated statistical analysis in R. Bioinformatics. 2016; 32:952–4.

18. Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012; 2:401–4.

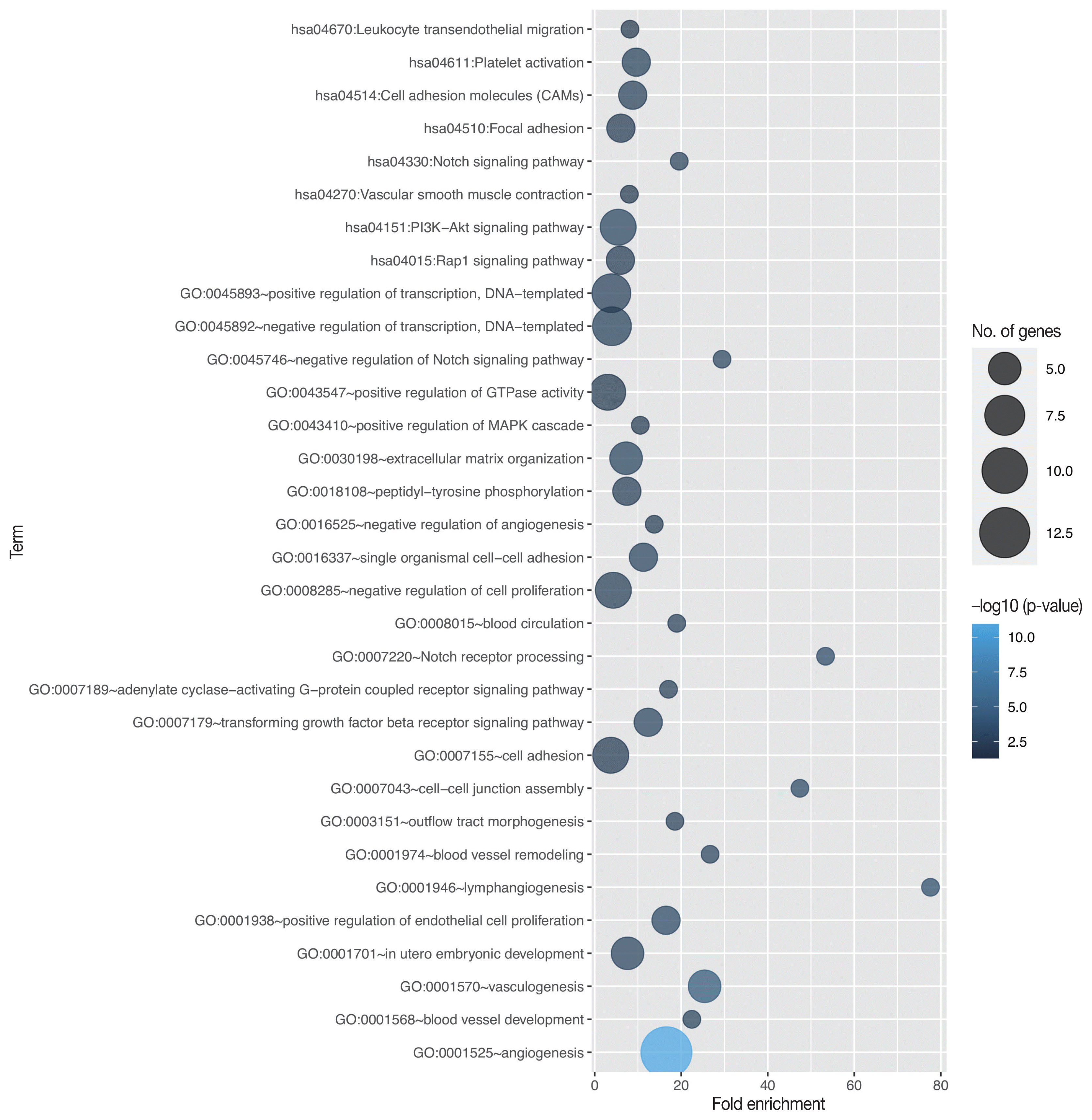
19. Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009; 37:1–13.

20. Louis DN, Perry A, Wesseling P, et al. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol. 2021; 23:1231–51.

21. Cheng Z, Dai Y, Pang Y, et al. High EGFL7 expression may predict poor prognosis in acute myeloid leukemia patients undergoing allogeneic hematopoietic stem cell transplantation. Cancer Biol Ther. 2019; 20:1314–8.
22. Papaioannou D, Shen C, Nicolet D, et al. Prognostic and biological significance of the proangiogenic factor EGFL7 in acute myeloid leukemia. Proc Natl Acad Sci U S A. 2017; 114:E4641–7.

23. Yang C, Wang YL, Sun D, Zhu XL, Li Z, Ni CF. Increased expression of epidermal growth factor-like domain-containing protein 7 is predictive of poor prognosis in patients with hepatocellular carcinoma. J Cancer Res Ther. 2018; 14:867–72.

24. Oh J, Park SH, Lee TS, Oh HK, Choi JH, Choi YS. High expression of epidermal growth factor-like domain 7 is correlated with poor differentiation and poor prognosis in patients with epithelial ovarian cancer. J Gynecol Oncol. 2014; 25:334–41.

25. Li Z, Xue TQ, Yang C, Wang YL, Zhu XL, Ni CF. EGFL7 promotes hepatocellular carcinoma cell proliferation and inhibits cell apoptosis through increasing CKS2 expression by activating Wnt/beta-catenin signaling. J Cell Biochem. 2018; 119:10327–37.
26. Liu Q, Zhang J, Gao H, et al. Role of EGFL7/EGFR-signaling pathway in migration and invasion of growth hormone-producing pituitary adenomas. Sci China Life Sci. 2018; 61:893–901.

27. Nichol D, Shawber C, Fitch MJ, et al. Impaired angiogenesis and altered Notch signaling in mice overexpressing endothelial Egfl7. Blood. 2010; 116:6133–43.

28. Bill M, Pathmanathan A, Karunasiri M, et al. EGFL7 antagonizes NOTCH signaling and represents a novel therapeutic target in acute myeloid leukemia. Clin Cancer Res. 2020; 26:669–78.

29. Tang H, Xiao WR, Liao YY, et al. EGFL7 silencing inactivates the Notch signaling pathway; enhancing cell apoptosis and suppressing cell proliferation in human cutaneous melanoma. Neoplasma. 2019; 66:187–96.

30. Schmidt M, De Maziere A, Smyczek T, et al. The role of Egfl7 in vascular morphogenesis. Novartis Found Symp. 2007; 283:18–28.
31. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011; 144:646–74.

32. Yeung S, Smyczek T, Cheng J, et al. Abstract 3295: inhibiting vascular morphogenesis in tumors: EGFL7 as a novel therapeutic target. Cancer Res. 2011; 71:3295.

33. Garcia-Carbonero R, van Cutsem E, Rivera F, et al. Randomized phase II trial of parsatuzumab (anti-EGFL7) or placebo in combination with FOLFOX and bevacizumab for first-line metastatic colorectal cancer. Oncologist. 2017; 22:375.

34. von Pawel J, Spigel DR, Ervin T, et al. Randomized phase II trial of parsatuzumab (anti-EGFL7) or placebo in combination with carboplatin, paclitaxel, and bevacizumab for first-line nonsquamous non-small cell lung cancer. Oncologist. 2018; 23:654.

35. Bicker F, Schmidt MH. EGFL7: a new player in homeostasis of the nervous system. Cell Cycle. 2010; 9:1263–9.

36. Shahcheraghi SH, Tchokonte-Nana V, Lotfi M, Lotfi M, Ghorbani A, Sadeghnia HR. Wnt/beta-catenin and PI3K/Akt/mTOR signaling pathways in glioblastoma: two main targets for drug design: a review. Curr Pharm Des. 2020; 26:1729–41.

37. Bos JL, de Rooij J, Reedquist KA. Rap1 signalling: adhering to new models. Nat Rev Mol Cell Biol. 2001; 2:369–77.

38. Han J, Lim CJ, Watanabe N, et al. Reconstructing and deconstructing agonist-induced activation of integrin alphaIIbbeta3. Curr Biol. 2006; 16:1796–806.
39. Sayyah J, Bartakova A, Nogal N, Quilliam LA, Stupack DG, Brown JH. The Ras-related protein, Rap1A, mediates thrombin-stimulated, integrin-dependent glioblastoma cell proliferation and tumor growth. J Biol Chem. 2014; 289:17689–98.