Abstract
Background and Purpose
To explore the causal relationships of elements of the exposome with ischemic stroke and its subtypes at the omics level and to provide evidence for stroke prevention.
Methods
We conducted a Mendelian randomization study between exposure and any ischemic stroke (AIS) and its subtypes (large-artery atherosclerotic disease [LAD], cardioembolic stroke [CE], and small vessel disease [SVD]). The exposure dataset was the UK Biobank involving 361,194 subjects, and the outcome dataset was the MEGASTROKE consortium including 52,000 participants.
Results
We found that higher blood pressure (BP) (systolic BP: odds ratio [OR], 1.02; 95% confidence interval [CI], 1.01 to 1.04; diastolic BP: OR, 1.03; 95% CI, 1.01 to 1.05; pulse pressure: OR, 1.03; 95% CI, 1.00 to 1.06), atrial fibrillation (OR, 1.18; 95% CI, 1.13 to 1.25), and diabetes (OR, 1.13; 95% CI, 1.07 to 1.18) were significantly associated with ischemic stroke. Importantly, higher education (OR, 0.69; 95% CI, 0.60 to 0.79) decreased the risk of ischemic stroke. Higher systolic BP (OR, 1.06; 95% CI, 1.02 to 1.10), pulse pressure (OR, 1.08; 95% CI, 1.02 to 1.14), diabetes (OR, 1.28; 95% CI, 1.13 to 1.45), and coronary artery disease (OR, 1.58; 95% CI, 1.25 to 2.00) could cause LAD. Atrial fibrillation could cause CE (OR, 1.90; 95% CI, 1.71 to 2.11). For SVD, higher systolic BP (OR, 1.04; 95% CI, 1.00 to 1.07), diastolic BP (OR, 1.06; 95% CI, 1.01 to 1.12), and diabetes (OR, 1.22; 95% CI, 1.10 to 1.36) were causal factors.
As one of the most devastating neurological diseases, stroke is a leading cause of mortality and adult disability worldwide, especially in low- and middle-income regions [1]. It accounts for 10% of disability-adjusted life-years lost and 5% of deaths annually [2]. With the increasing global burden of stroke, identifying the underlying risks and protective factors is crucial for stroke prevention. Previous observational studies have reported that 90% of strokes are attributed to modifiable risk factors [3]. However, observational data are limited by confounding and reverse causality, leading to limited power to identify causal associations. The method of Mendelian randomization (MR) has become a powerful tool for investigating the causal relationships between risk factors and disease using observational data [4]. MR studies use genetic information as instrumental variables to implement causal relationships, and it can be regarded as analogous to a randomized controlled study [5].
The concept of exposome was first proposed to set a high-throughput method to elucidate the relationships between all exposures and a disease [6]. Exposome consist of the entire set of environmental exposures, ranging from individual-level (e.g., education, cigarette smoking, exercise, and hypertension) to exogenous-level exposures (e.g., air pollution and socioeconomic status) [7]. In parallel to -omics (e.g., genomics, metabolomics), exposome superseded the characteristics of exposures “one by one.” Therefore, the stroke exposome concept can be used to comprehensively detect the causal factors of stroke at the omics level, while conventional epidemiological risk factor studies focusing on one or several exposures at a time may miss some causal and preventive factors. We conducted exposome-wide association studies (ExWAS) to evaluate the potential causal effects of multiple exposures on stroke via the MR method based on the concept of exposome [8].
In this study, we aimed to assess the causal associations between exposure (including exogenous and endogenous factors) and ischemic stroke, as well as its subtypes (i.e., large-artery atherosclerotic disease [LAD], cardioembolic stroke [CE], and small vessel disease [SVD]). We conducted an ExWAS using the MR method to detect possible exposure-stroke associations in the hope of preventing stroke. To the best of our knowledge, this is the first study to use the MR method to evaluate the causal relationship between exposure and stroke.
Summary-level data were obtained from 4,587 genome-wide association studies (GWAS) analyzed by Neale Lab (http://www.nealelab.is/uk-biobank) on various exposures conducted in 361,194 participants from the UK Biobank. The UK Biobank is a prospective cohort study with deep genetic data and broad individual phenotypic and health-related data [9]. The least-squares linear model was used to test the associations of all exposures with sex and the first 10 principal components as covariates.
Based on the previous conception of exposome, we classified all the exposures into three major domains [10-13], exogenous macro-level domain, exogenous individual domain, and endogenous domain. Overall, 76, 1,306, and 1,521 exposures were classified into the exogenous macro-level domain, exogenous individual domain, and endogenous domain, respectively. Single nucleotide polymorphisms (SNPs) associated with exposures in GWAS analyses with a P<1×10-6 were defined as instrument variables. SNPs which correlated with the top SNPs at r2 >0.001 were excluded for independence. We used the following criteria for exposure GWAS to be included in this study: (1) the exposure GWAS identified SNPs with a P<1×10-6 [14]; (2) cases of binary exposures >250, or samples with continuous exposures >250 [15]; (3) instrument SNPs >3. The exposures violated the basic assumption of the MR study, and exposures with unclear definitions were excluded. A flow chart is showed in Figure 1.
Summary statistics of stroke and stroke subtypes were drawn from a recent large-scale meta-analysis of GWAS (MEGASTROKE) confined to European populations (40,585 cases; 406,111 controls) [16]. Specifically, any ischemic stroke (AIS), LAD, CE, and SVD were screened for potential casual exposures. A detailed description of the participants and study design of MEGASTROKE were provided in the original study [16].
Harmonization was conducted to ensure that each SNP of exposure and stroke corresponded to the same strand. Primarily, we used the inverse-variance weighted (IVW) method to identify the relationship between the components of the exposome and stroke and its subtypes. Furthermore, we used the MR Egger and weight media method to estimate horizontal pleiotropy [24,25]. We used the F-statistic to evaluate the strength of the instrumental variables, and we estimated the power of this MR with a false positive rate α=0.05 [26,27]. In the screening stage, Bonferroni-adjusted P<0.05/n was considered to be statistically significant. In the validation stage, statistical significance was set at P<0.05. All analyses were conducted in the environment of R version 3.5.1 (R Foundation for Statistical Computing, Vienna, Austria), and the TwoSampleMR package in R was applied to perform MR analyses [28,29].
Overall, 4,587 exposures were tested for causal associations with ischemic stroke and stroke subtypes. Prior to the association analysis, all exposures underwent stringent quality control. Subsequently, 2,160, 2,114, 2,120, and 2,119 exposures survived the filtering process for AIS, LAD, CE, and SVD, respectively. Statistical significance (IVW Bonferroni-adjusted P-value) was reached in nine exposures for AIS, six exposures in LAD, three exposures in CE, and five exposures in SVD (Figure 2).
A list of all the exposures with a significant association with AIS or stroke subtype is presented in Table 1. Hypertension and diabetes were diagnosed by physicians or self-reported. Three categories of exposure were significantly associated with AIS: diseases (hypertension, P=1.49E-25; diabetes, P=5.29E-08; atrial fibrillation [AF] and flutter, P=7.25E-09; cardiac arrhythmia and comorbidities of chronic obstructive pulmonary disease [COPD], P=3.15E-06), blood pressure (BP) (systolic BP [SBP], P=3.38E-11; diastolic BP [DBP], P=8.28E-08), and education (college or university degree, P=1.81E-06). Similarly, LAD was also potentially caused by three categories of exposure, including diseases (hypertension, P=2.47E-12; coronary atherosclerosis, P=8.96E-06), family history (family history of diabetes, P=4.80E-06), and BP (SBP, P=2.48E-10). Only exposure to disease showed associations with CE, including AF/flutter (P=4.91E-10) and hypertension (P=5.28E-09). Similar to LAD, SVD was potentially caused by diseases (diabetes, P=2.47E-06; hypertension, P=4.60E-14), high BP (DBP, P=6.27E-06), and family history (family history of hypertension, P=1.57E-07). All F stat and R2 indicated high strength of this MR. We did not identify directional pleiotropy or outliers in the associations between the significant exposures and stroke (Supplementary Figures 1-23).
To validate the exposures showing a causal relationship with AIS or stroke subtype, we chose the most recent, large-scale GWAS of the identified exposures and further tested the causal relationships between the exposures and stroke/stroke subtypes in two-sample MR analysis. We replicated exposures with causal effects on stroke or stroke subtypes (Figure 3) [17-23]. Genetically determined higher BP levels (SBP: odds ratio [OR], 1.02; 95% confidence interval [CI], 1.01 to 1.04; IVW P=0.0061; DBP: OR, 1.03; 95% CI, 1.01 to 1.05; IVW P=0.013; pulse pressure [PP]: OR, 1.03; 95% CI, 1.00 to 1.06; IVW P=0.023), AF (OR, 1.18; 95% CI, 1.13 to 1.25; IVW P=3.76E-11), and diabetes (OR, 1.13; 95% CI, 1.07 to 1.18; IVW P=2.80E-06) were significantly associated with a higher risk of ischemic stroke, whereas higher education (OR, 0.69; 95% CI, 0.60 to 0.79; IVW P=8.82E-08) was associated with a lower risk of ischemic stroke. COPD showed no significant causal association with ischemic stroke (OR, 0.76; 95% CI, 0.60 to 0.79; IVW, P=0.052).
Specifically, higher SBP (OR, 1.06; 95% CI, 1.02 to 1.10; IVW P=0.0052), higher PP (OR, 1.08; 95% CI, 1.02 to 1.14; IVW P=0.0095), diabetes (OR, 1.28; 95% CI, 1.13 to 1.45; IVW P=8.54E-05), and coronary artery disease (CAD; OR, 1.58; 95% CI, 1.25 to 2.00; IVW P=0.00012) increased the risk of LAD, whereas higher DBP was not significantly associated with LAD (OR, 1.06; 95% CI, 0.99 to 1.12; IVW P=0.088). Only AF was replicated as a causal factor for CE (OR, 1.90; 95% CI, 1.71 to 2.11; IVW P=4.33E-34). Similar to LAD, the replicated causal exposures of SVD included higher SBP (OR, 1.04; 95% CI, 1.00 to 1.07; IVW P=0.044), higher DBP (OR, 1.06; 95% CI, 1.01 to 1.12; IVW P=0.0012), and diabetes (OR, 1.22; 95% CI, 1.10 to 1.36; IVW P=0.00024); however, PP showed no significant causal association with SVD (OR, 0.97; 95% CI, 1.03 to 1.08; IVW P=0.37). Results of the MR sensitivity analyses were in accordance with the primary results. No evidence of directional pleiotropy or outlier association was detected (Supplementary Figures 24-43).
To elucidate the preventive effect of higher education, we further investigated various aspects of education, including educational attainment, years of education, highest math classes taken, and cognitive performance (Table 2). After correction for multiple hypotheses testing, we found that genetically determined higher educational attainment and longer time of education could significantly decrease the risk of stroke (OR, 0.65; 95% CI, 0.57 to 0.74; IVW P=3.63E-11; and OR, 0.66; 95% CI, 0.59 to 0.75; IVW P=3.25E-11, respectively). Taking math classes and better cognitive performance were also associated with a lower risk of stroke (OR, 0.80; 95% CI, 0.71 to 0.90; IVW P=0.00029). We did not detect evidence of pleiotropy, and the other MR methods indicated consistent estimates of the effect (Supplementary Figures 44-46).
In this MR study of exposure and stroke, we identified traditional causal mediators for ischemic stroke (higher SBP, DBP, PP, AF, and diabetes), LAD (higher SBP, PP, diabetes, and CAD), CE (AF), and SVD (higher SBP, DBP, and diabetes). Of note, we found education attainment, length of education, math classes taken, and cognitive performance as novel causal exposures of ischemic stroke.
Our study indicates that education is a novel protective causal factor for AIS. Whether education was associated with stroke was inconclusive before our current study. A large Australian prospective cohort study reported that low education was associated with increased stroke risk (adjusted hazard ratio [HR], 1.41; 95% CI, 1.16 to 1.71; and HR, 1.25; 95% CI, 1.07 to 1.46, for women and men, respectively) [30]. Similarly, a large cohort study following patients up for 26 years indicated that educational attainment had an inverse dose-dependent relationship with cerebrovascular disease. However, the first National Health and Nutrition Examination Survey, an epidemiological longitudinal study, suggested that the association between educational attainment and stroke was not statistically significant [31]. These observational studies did not provide solid evidence for the stroke prevention effect of higher education. In this study, we used a genetic instrument to simulate a randomized controlled study that attempted to reveal the relationship between education and stroke. Consistent with previous studies, higher education attainment was found to be associated with a lower risk of stroke. Thus, strategies that diminish education inequalities are of great importance for stroke prevention.
The causal risk factors identified by this exposome MR study were consistent with previously well-known traditional risk factors for stroke (hypertension, diabetes, coronary artery atherosclerosis, and AF) [32]. Furthermore, the casual exposures of its subtypes were also consistent with conventional conception, and thus validated our exposome MR method.
Our investigation has methodological strengths. First, we used a high-throughput, data-driven approach to screen potential causal exposures at the omics level. We evaluated as many exposures as possible, leveraging the large sample size and enriched information from the UK Biobank. Compared with conventional observational studies or randomized clinical trials that can only appraise one or several risk factors, we evaluated thousands of exposures in one study. Second, the MR study could be considered as a randomization study. Leveraging genetics, two-sample MR could be used to infer the causality of exposures and outcomes. Furthermore, the identified exposures through the first screening step were replicated by the most recent and largest GWAS, providing validation for the exposome MR study.
This study has some limitations. First, the association model of GWAS of exposures was a least-squares linear model, regardless of whether the exposure variables were continuous or binary. The binary traits were better suited for the logistic model, and the linear model may create biases in the beta coefficients. Thus, we noticed that the estimation of associations strongly deviated to binary or ordinal variables. However, dealing with large-scale data with different regression models would result in unnecessary complexity. Furthermore, the tendency of the causal relationship could also be detected using the linear model. Importantly, our replication study confirmed the exposome MR results. Second, we detected causal exposures of stroke and stroke subtypes. However, the dosage effects of risk exposures on stroke have not been investigated. The MR method focused on explaining the causal relationship between exposure and outcome. The dosage effects of exposure should be investigated further. Additionally, this study detected causal exposure by using MR between the exposome and stroke, but we did not weigh multiple exposures using this method. Weighing the risks of the detected factors is required in the future. Further research on the polygenic risk score is needed to explore the relationship between genetics, environmental exposures, and outcomes in a large longitudinal cohort. Finally, we identified several modifiable risk factors for stroke and stroke subtypes. Our study provides an interventional treatment target. Optimal medication and drug compliance are of great importance in controlling these risk factors.
We screened the causal exposures of stroke at the omics level using a MR study. Traditional risk factors such as hypertension, diabetes, CAD, and AF were confirmed as causative relationships that contributed to all stroke and stroke subtypes. Educational attainment can reduce the incidence of AIS. Our findings suggest that improving the education level and managing modifiable causal factors are essential for stroke prevention.
Supplementary materials
Supplementary materials related to this article can be found online at https://doi.org/10.5853/jos.2021.01340.
Supplementary Figure 1.
The leave-one-out analysis, single nucleotide polymorphism (SNP) analysis, sensitivity analysis, and funnel plot for self-reported: hypertension and any ischemic stroke (AIS). (A) Leave-one-out analysis, (B) forest plot, (C) comparison of results using different Mendelian randomization (MR) methods, (D) funnel plot of inverse-variance weighted (IVW) and MR-Egger regression. SE, standard error.
Supplementary Figure 2.
The leave-one-out analysis, single nucleotide polymorphism (SNP) analysis, sensitivity analysis, and funnel plot for hypertension and any ischemic stroke (AIS). (A) Leave-one-out analysis, (B) forest plot, (C) comparison of results using different Mendelian randomization (MR) methods, (D) funnel plot of inverse-variance weighted (IVW) and MR-Egger regression. SE, standard error.
Supplementary Figure 3.
The leave-one-out analysis, single nucleotide polymorphism (SNP) analysis, sensitivity analysis, and funnel plot for systolic blood pressure and any ischemic stroke (AIS). (A) Leave-one-out analysis, (B) forest plot, (C) comparison of results using different Mendelian randomization (MR) methods, (D) funnel plot of inverse-variance weighted (IVW) and MR-Egger regression. SE, standard error.
Supplementary Figure 4.
The leave-one-out analysis, single nucleotide polymorphism (SNP) analysis, sensitivity analysis, and funnel plot for self-reported: diabetes mellitus and any ischemic stroke (AIS). (A) Leave-one-out analysis, (B) forest plot, (C) comparison of results using different Mendelian randomization (MR) methods, (D) funnel plot of inverse-variance weighted (IVW) and MR-Egger regression. SE, standard error.
Supplementary Figure 5.
The leave-one-out analysis, single nucleotide polymorphism (SNP) analysis, sensitivity analysis, and funnel plot for atrial fibrillation/flutter and any ischemic stroke (AIS). (A) Leave-one-out analysis, (B) forest plot, (C) comparison of results using different Mendelian randomization (MR) methods, (D) funnel plot of inverse-variance weighted (IVW) and MR-Egger regression. SE, standard error.
Supplementary Figure 6.
The leave-one-out analysis, single nucleotide polymorphism (SNP) analysis, sensitivity analysis, and funnel plot for diabetes mellitus and any ischemic stroke (AIS). (A) Leave-one-out analysis, (B) forest plot, (C) comparison of results using different Mendelian randomization (MR) methods, (D) funnel plot of inverse-variance weighted (IVW) and MR-Egger regression. SE, standard error.
Supplementary Figure 7.
The leave-one-out analysis, single nucleotide polymorphism (SNP) analysis, sensitivity analysis, and funnel plot for diastolic blood pressure and any ischemic stroke (AIS). (A) Leave-one-out analysis, (B) forest plot, (C) comparison of results using different Mendelian randomization (MR) methods, (D) funnel plot of inverse-variance weighted (IVW) and MR-Egger regression. SE, standard error.
Supplementary Figure 8.
The leave-one-out analysis, single nucleotide polymorphism (SNP) analysis, sensitivity analysis, and funnel plot for College or University degree and any ischemic stroke (AIS). (A) Leave-one-out analysis, (B) forest plot, (C) comparison of results using different Mendelian randomization (MR) methods, (D) funnel plot of inverse-variance weighted (IVW) and MR-Egger regression. SE, standard error.
Supplementary Figure 9.
The leave-one-out analysis, single nucleotide polymorphism (SNP) analysis, sensitivity analysis, and funnel plot for cardiac arrhythmias, chronic obstructive pulmonary disease co-morbidities and any ischemic stroke (AIS). (A) Leave-one-out analysis, (B) forest plot, (C) comparison of results using different Mendelian randomization (MR) methods, (D) funnel plot of inverse-variance weighted (IVW) and MR-Egger regression. SE, standard error.
Supplementary Figure 10.
The leave-one-out analysis, single nucleotide polymorphism (SNP) analysis, sensitivity analysis, and funnel plot for hypertension and large-artery atherosclerotic stroke (LAS). (A) Leave-one-out analysis, (B) forest plot, (C) comparison of results using different Mendelian randomization (MR) methods, (D) funnel plot of inverse-variance weighted (IVW) and MR-Egger regression. SE, standard error.
Supplementary Figure 11.
The leave-one-out analysis, single nucleotide polymorphism (SNP) analysis, sensitivity analysis, and funnel plot for self-reported: hypertension and large-artery atherosclerotic stroke (LAS). (A) Leave-one-out analysis, (B) forest plot, (C) comparison of results using different Mendelian randomization (MR) methods, (D) funnel plot of inverse-variance weighted (IVW) and MR-Egger regression. SE, standard error.
Supplementary Figure 12.
The leave-one-out analysis, single nucleotide polymorphism (SNP) analysis, sensitivity analysis, and funnel plot for systolic blood pressure and large-artery atherosclerotic stroke (LAS). (A) Leave-one-out analysis, (B) forest plot, (C) comparison of results using different Mendelian randomization (MR) methods, (D) funnel plot of inverse-variance weighted (IVW) and MR-Egger regression. SE, standard error.
Supplementary Figure 13.
The leave-one-out analysis, single nucleotide polymorphism (SNP) analysis, sensitivity analysis, and funnel plot for diabetes mellitus and large-artery atherosclerotic stroke (LAS). (A) Leave-one-out analysis, (B) forest plot, (C) comparison of results using different Mendelian randomization (MR) methods, (D) funnel plot of inverse-variance weighted (IVW) and MR-Egger regression. SE, standard error.
Supplementary Figure 14.
The leave-one-out analysis, single nucleotide polymorphism (SNP) analysis, sensitivity analysis and funnel plot for illnesses of father: diabetes mellitus and large-artery atherosclerotic stroke (LAS). (A) Leave-one-out analysis, (B) forest plot, (C) comparison of results using different Mendelian randomization (MR) methods, (D) funnel plot of inverse-variance weighted (IVW) and MR-Egger regression. SE, standard error.
Supplementary Figure 15.
The leave-one-out analysis, single nucleotide polymorphism (SNP) analysis, sensitivity analysis, and funnel plot for coronary atherosclerosis and large-artery atherosclerotic stroke (LAS). (A) Leave-one-out analysis, (B) forest plot, (C) comparison of results using different Mendelian randomization (MR) methods, (D) funnel plot of inverse-variance weighted (IVW) and MR-Egger regression. SE, standard error.
Supplementary Figure 16.
The leave-one-out analysis, single nucleotide polymorphism (SNP) analysis, sensitivity analysis, and funnel plot for atrial fibrillation/flutter and cardio-embolic stroke (CES). (A) Leave-one-out analysis, (B) forest plot, (C) comparison of results using different Mendelian randomization (MR) methods, (D) funnel plot of inverse-variance weighted (IVW) and MR-Egger regression. SE, standard error.
Supplementary Figure 17.
The leave-one-out analysis, single nucleotide polymorphism (SNP) analysis, sensitivity analysis, and funnel plot for self-reported: hypertension and cardio-embolic stroke (CES). (A) Leave-one-out analysis, (B) forest plot, (C) comparison of results using different Mendelian randomization (MR) methods, (D) funnel plot of inverse-variance weighted (IVW) and MR-Egger regression. SE, standard error.
Supplementary Figure 18.
The leave-one-out analysis, single nucleotide polymorphism (SNP) analysis, sensitivity analysis, and funnel plot for hypertension and cardio-embolic stroke (CES). (A) Leave-one-out analysis, (B) forest plot, (C) comparison of results using different Mendelian randomization (MR) methods, (D) funnel plot of inverse-variance weighted (IVW) and MR-Egger regression. SE, standard error.
Supplementary Figure 19.
The leave-one-out analysis, single nucleotide polymorphism (SNP) analysis, sensitivity analysis, and funnel plot for diastolic blood pressure and small-vessel stroke (SVS). (A) Leave-one-out analysis, (B) forest plot, (C) comparison of results using different Mendelian randomization (MR) methods, (D) funnel plot of inverse-variance weighted (IVW) and MR-Egger regression. SE, standard error.
Supplementary Figure 20.
The leave-one-out analysis, single nucleotide polymorphism (SNP) analysis, sensitivity analysis, and funnel plot for illnesses of siblings: hypertension and small-vessel stroke (SVS). (A) Leave-one-out analysis, (B) forest plot, (C) comparison of results using different Mendelian randomization (MR) methods, (D) funnel plot of inverse-variance weighted (IVW) and MR-Egger regression. SE, standard error.
Supplementary Figure 21.
The leave-one-out analysis, single nucleotide polymorphism (SNP) analysis, sensitivity analysis, and funnel plot for self-reported: diabetes mellitus and small-vessel stroke (SVS). (A) Leave-one-out analysis, (B) forest plot, (C) comparison of results using different Mendelian randomization (MR) methods, (D) funnel plot of inverse-variance weighted (IVW) and MR-Egger regression. SE, standard error.
Supplementary Figure 22.
The leave-one-out analysis, single nucleotide polymorphism (SNP) analysis, sensitivity analysis, and funnel plot for self-reported: hypertension and small-vessel stroke (SVS). (A) Leave-one-out analysis, (B) forest plot, (C) comparison of results using different Mendelian randomization (MR) methods, (D) funnel plot of inverse-variance weighted (IVW) and MR-Egger regression. SE, standard error.
Supplementary Figure 23.
The leave-one-out analysis, single nucleotide polymorphism (SNP) analysis, sensitivity analysis, and funnel plot for hypertension and small-vessel stroke (SVS). (A) Leave-one-out analysis, (B) forest plot, (C) comparison of results using different Mendelian randomization (MR) methods, (D) funnel plot of inverse-variance weighted (IVW) and MR-Egger regression. SE, standard error.
Supplementary Figure 24.
The leave-one-out analysis, single nucleotide polymorphism (SNP) analysis, sensitivity analysis, and funnel plot for systolic blood pressure and any ischemic stroke (AIS). (A) Leave-one-out analysis, (B) forest plot, (C) comparison of results using different Mendelian randomization (MR) methods, (D) funnel plot of inverse-variance weighted (IVW) and MR-Egger regression. SE, standard error.
Supplementary Figure 25.
The leave-one-out analysis, single nucleotide polymorphism (SNP) analysis, sensitivity analysis, and funnel plot for diastolic blood pressure and any ischemic stroke (AIS). (A) Leave-one-out analysis, (B) forest plot, (C) comparison of results using different Mendelian randomization (MR) methods, (D) funnel plot of inverse-variance weighted (IVW) and MR-Egger regression. SE, standard error.
Supplementary Figure 26.
The leave-one-out analysis, single nucleotide polymorphism (SNP) analysis, sensitivity analysis, and funnel plot for pulse pressure and any ischemic stroke (AIS). (A) Leave-one-out analysis, (B) forest plot, (C) comparison of results using different Mendelian randomization (MR) methods, (D) funnel plot of inverse-variance weighted (IVW) and MR-Egger regression. SE, standard error.
Supplementary Figure 27.
The leave-one-out analysis, single nucleotide polymorphism (SNP) analysis, sensitivity analysis, and funnel plot for atrial fibrillation and any ischemic stroke (AIS). (A) Leave-one-out analysis, (B) forest plot, (C) comparison of results using different Mendelian randomization (MR) methods, (D) funnel plot of inverse-variance weighted (IVW) and MR-Egger regression. SE, standard error.
Supplementary Figure 28.
The leave-one-out analysis, single nucleotide polymorphism (SNP) analysis, sensitivity analysis, and funnel plot for diabetes mellitus and any ischemic stroke (AIS). (A) Leave-one-out analysis, (B) forest plot, (C) comparison of results using different Mendelian randomization (MR) methods, (D) funnel plot of inverse-variance weighted (IVW) and MR-Egger regression. SE, standard error.
Supplementary Figure 29.
The leave-one-out analysis, single nucleotide polymorphism (SNP) analysis, sensitivity analysis, and funnel plot for education attainment and any ischemic stroke (AIS). (A) Leave-one-out analysis, (B) forest plot, (C) comparison of results using different Mendelian randomization (MR) methods, (D) funnel plot of inverse-variance weighted (IVW) and MR-Egger regression. SE, standard error.
Supplementary Figure 30.
The leave-one-out analysis, single nucleotide polymorphism (SNP) analysis, sensitivity analysis, and funnel plot for lung fuction and any ischemic stroke (AIS). (A) Leave-one-out analysis, (B) forest plot, (C) comparison of results using different Mendelian randomization (MR) methods, (D) funnel plot of inverse-variance weighted (IVW) and MR-Egger regression. FEV1, forced expiratory volume in 1 second; FVC, forced vital capacity; SE, standard error.
Supplementary Figure 31.
The leave-one-out analysis, single nucleotide polymorphism (SNP) analysis, sensitivity analysis, and funnel plot for systolic blood pressure and large-artery atherosclerotic stroke (LAS). (A) Leave-one-out analysis, (B) forest plot, (C) comparison of results using different Mendelian randomization (MR) methods, (D) funnel plot of inverse-variance weighted (IVW) and MR-Egger regression. SE, standard error.
Supplementary Figure 32.
The leave-one-out analysis, single nucleotide polymorphism (SNP) analysis, sensitivity analysis, and funnel plot for diastolic blood pressure and large-artery atherosclerotic stroke (LAS). (A) Leave-one-out analysis, (B) forest plot, (C) comparison of results using different Mendelian randomization (MR) methods, (D) funnel plot of inverse-variance weighted (IVW) and MR-Egger regression. SE, standard error.
Supplementary Figure 33.
The leave-one-out analysis, single nucleotide polymorphism (SNP) analysis, sensitivity analysis, and funnel plot for pulse pressure and large-artery atherosclerotic stroke (LAS). (A) Leave-one-out analysis, (B) forest plot, (C) comparison of results using different Mendelian randomization (MR) methods, (D) funnel plot of inverse-variance weighted (IVW) and MR-Egger regression. SE, standard error.
Supplementary Figure 34.
The leave-one-out analysis, single nucleotide polymorphism (SNP) analysis, sensitivity analysis, and funnel plot for diabetes mellitus and large-artery atherosclerotic stroke (LAS). (A) Leave-one-out analysis, (B) forest plot, (C) comparison of results using different Mendelian randomization (MR) methods, (D) funnel plot of inverse-variance weighted (IVW) and MR-Egger regression. SE, standard error.
Supplementary Figure 35.
The leave-one-out analysis, single nucleotide polymorphism (SNP) analysis, sensitivity analysis, and funnel plot for coronary heart disease (CHD) and large-artery atherosclerotic stroke (LAS). (A) Leave-one-out analysis, (B) forest plot, (C) comparison of results using different Mendelian randomization (MR) methods, (D) funnel plot of inverse-variance weighted (IVW) and MR-Egger regression. SE, standard error.
Supplementary Figure 36.
The leave-one-out analysis, single nucleotide polymorphism (SNP) analysis, sensitivity analysis, and funnel plot for systolic blood pressure and cardio-embolic stroke (CES). (A) Leave-one-out analysis, (B) forest plot, (C) comparison of results using different Mendelian randomization (MR) methods, (D) funnel plot of inverse-variance weighted (IVW) and MR-Egger regression. SE, standard error.
Supplementary Figure 37.
The leave-one-out analysis, single nucleotide polymorphism (SNP) analysis, sensitivity analysis, and funnel plot for diastolic blood pressure and cardio-embolic stroke (CES). (A) Leave-one-out analysis, (B) forest plot, (C) comparison of results using different Mendelian randomization (MR) methods, (D) funnel plot of inverse-variance weighted (IVW) and MR-Egger regression. SE, standard error.
Supplementary Figure 38.
The leave-one-out analysis, single nucleotide polymorphism (SNP) analysis, sensitivity analysis, and funnel plot for pulse pressure and cardio-embolic stroke (CES). (A) Leave-one-out analysis, (B) forest plot, (C) comparison of results using different Mendelian randomization (MR) methods, (D) funnel plot of inverse-variance weighted (IVW) and MR-Egger regression. SE, standard error.
Supplementary Figure 39.
The leave-one-out analysis, single nucleotide polymorphism (SNP) analysis, sensitivity analysis, and funnel plot for atrial fibrillation and cardio-embolic stroke (CES). (A) Leave-one-out analysis, (B) forest plot, (C) comparison of results using different Mendelian randomization (MR) methods, (D) funnel plot of inverse-variance weighted (IVW) and MR-Egger regression. SE, standard error.
Supplementary Figure 40.
The leave-one-out analysis, single nucleotide polymorphism (SNP) analysis, sensitivity analysis, and funnel plot for systolic blood pressure and small-vessel stroke (SVS). (A) Leave-one-out analysis, (B) forest plot, (C) comparison of results using different Mendelian randomization (MR) methods, (D) funnel plot of inverse-variance weighted (IVW) and MR-Egger regression. SE, standard error.
Supplementary Figure 41.
The leave-one-out analysis, single nucleotide polymorphism (SNP) analysis, sensitivity analysis, and funnel plot for diastolic blood pressure and small-vessel stroke (SVS). (A) Leave-one-out analysis, (B) forest plot, (C) comparison of results using different Mendelian randomization (MR) methods, (D) funnel plot of inverse-variance weighted (IVW) and MR-Egger regression. SE, standard error.
Supplementary Figure 42.
The leave-one-out analysis, single nucleotide polymorphism (SNP) analysis, sensitivity analysis, and funnel plot for pulse pressure and small-vessel stroke (SVS). (A) Leave-one-out analysis, (B) forest plot, (C) comparison of results using different Mendelian randomization (MR) methods, (D) funnel plot of inverse-variance weighted (IVW) and MR-Egger regression. SE, standard error.
Supplementary Figure 43.
The leave-one-out analysis, single nucleotide polymorphism (SNP) analysis, sensitivity analysis, and funnel plot for diabetes mellitus and small-vessel stroke (SVS). (A) Leave-one-out analysis, (B) forest plot, (C) comparison of results using different Mendelian randomization (MR) methods, (D) funnel plot of inverse-variance weighted (IVW) and MR-Egger regression. SE, standard error.
Supplementary Figure 44.
The leave-one-out analysis, single nucleotide polymorphism (SNP) analysis, sensitivity analysis, and funnel plot for educational attainment (years of education) (unit increase) and any ischemic stroke (AIS). (A) Leave-one-out analysis, (B) forest plot, (C) comparison of results using different Mendelian randomization (MR) methods, (D) funnel plot of inverse-variance weighted (IVW) and MR-Egger regression. SE, standard error.
Supplementary Figure 45.
The leave-one-out analysis, single nucleotide polymorphism (SNP) analysis, sensitivity analysis, and funnel plot for highest math class taken and any ischemic stroke (AIS). (A) Leave-one-out analysis, (B) forest plot, (C) comparison of results using different Mendelian randomization (MR) methods, (D) funnel plot of inverse-variance weighted (IVW) and MR-Egger regression. SE, standard error.
Supplementary Figure 46.
The leave-one-out analysis, single nucleotide polymorphism (SNP) analysis, sensitivity analysis and funnel plot for cognitive performance (MTAG) (unit increase) and any ischemic stroke (AIS). (A) Leave-one-out analysis, (B) forest plot, (C) comparison of results using different Mendelian randomization (MR) methods, (D) funnel plot of inverse-variance weighted (IVW) and MR-Egger regression. SE, standard error.
ACKNOWLEDGMENTS
This work was made possible by the generous sharing of GWAS summary statistics. We thank Neale Lab for offering the GWAS of the exposure data. We also thank the UK Biobank, METASTROKE/MEGASTROKE Working Group (https://www.strokegenetics.org/node/317) for providing summary statistics for these analyses.
This study was supported by grants from the National Natural Science Foundation of China (91849126), the National Key R&D Program of China (2018YFC1314700), Shanghai Municipal Science and Technology Major Project (No.2018SHZDZX01), Zhangjiang Lab, Tianqiao and Chrissy Chen Institute, and the State Key Laboratory of Neurobiology and Frontiers Center for Brain Science of the Ministry of Education, Fudan University.
References
1. O’Donnell MJ, Chin SL, Rangarajan S, Xavier D, Liu L, Zhang H, et al. Global and regional effects of potentially modifiable risk factors associated with acute stroke in 32 countries (INTERSTROKE): a case-control study. Lancet. 2016; 388:761–775.

2. GBD 2016 Lifetime Risk of Stroke Collaborators, Feigin VL, Nguyen G, Cercy K, Johnson CO, Alam T, et al. Global, regional, and country-specific lifetime risks of stroke, 1990 and 2016. N Engl J Med. 2018; 379:2429–2437.

3. Feigin VL, Roth GA, Naghavi M, Parmar P, Krishnamurthi R, Chugh S, et al. Global burden of stroke and risk factors in 188 countries, during 1990-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet Neurol. 2016; 15:913–924.

4. Davies NM, Holmes MV, Davey Smith G. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ. 2018; 362:k601.

5. Zheng J, Baird D, Borges MC, Bowden J, Hemani G, Haycock P, et al. Recent developments in Mendelian randomization studies. Curr Epidemiol Rep. 2017; 4:330–345.

6. Wild CP. Complementing the genome with an “exposome”: the outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol Biomarkers Prev. 2005; 14:1847–1850.

7. Niedzwiecki MM, Walker DI, Vermeulen R, Chadeau-Hyam M, Jones DP, Miller GW. The exposome: molecules to populations. Annu Rev Pharmacol Toxicol. 2019; 59:107–127.

8. Nieuwenhuijsen MJ, Agier L, Basagaña X, Urquiza J, Tamayo-Uria I, Giorgis-Allemand L, et al. Influence of the urban exposome on birth weight. Environ Health Perspect. 2019; 127:47007.

9. Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018; 562:203–209.

10. Finch CE, Kulminski AM. The Alzheimer’s disease exposome. Alzheimers Dement. 2019; 15:1123–1132.

12. Rappaport SM. Implications of the exposome for exposure science. J Expo Sci Environ Epidemiol. 2011; 21:5–9.

13. Vrijheid M. The exposome: a new paradigm to study the impact of environment on health. Thorax. 2014; 69:876–878.

14. Doherty A, Smith-Byrne K, Ferreira T, Holmes MV, Holmes C, Pulit SL, et al. GWAS identifies 14 loci for device-measured physical activity and sleep duration. Nat Commun. 2018; 9:5257.

15. Noyce AJ, Bandres-Ciga S, Kim J, Heilbron K, Kia D, Hemani G, et al. The Parkinson’s disease Mendelian randomization research portal. Mov Disord. 2019; 34:1864–1872.

16. Malik R, Chauhan G, Traylor M, Sargurupremraj M, Okada Y, Mishra A, et al. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat Genet. 2018; 50:524–537.
17. Evangelou E, Warren HR, Mosen-Ansorena D, Mifsud B, Pazoki R, Gao H, et al. Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat Genet. 2018; 50:1412–1425.
18. Ellinor PT, Lunetta KL, Albert CM, Glazer NL, Ritchie MD, Smith AV, et al. Meta-analysis identifies six new susceptibility loci for atrial fibrillation. Nat Genet. 2012; 44:670–675.
19. Xue A, Wu Y, Zhu Z, Zhang F, Kemper KE, Zheng Z, et al. Genome-wide association analyses identify 143 risk variants and putative regulatory mechanisms for type 2 diabetes. Nat Commun. 2018; 9:2941.

20. Lee JJ, Wedow R, Okbay A, Kong E, Maghzian O, Zacher M, et al. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat Genet. 2018; 50:1112–1121.
21. Wain LV, Shrine N, Artigas MS, Erzurumluoglu AM, Noyvert B, Bossini-Castillo L, et al. Genome-wide association analyses for lung function and chronic obstructive pulmonary disease identify new loci and potential druggable targets. Nat Genet. 2017; 49:416–425.
22. Onengut-Gumuscu S, Chen WM, Burren O, Cooper NJ, Quinlan AR, Mychaleckyj JC, et al. Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat Genet. 2015; 47:381–386.

23. Nelson CP, Goel A, Butterworth AS, Kanoni S, Webb TR, Marouli E, et al. Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat Genet. 2017; 49:1385–1391.

24. Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016; 40:304–314.

25. Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015; 44:512–525.

26. Pierce BL, Ahsan H, Vanderweele TJ. Power and instrument strength requirements for Mendelian randomization studies using multiple genetic variants. Int J Epidemiol. 2011; 40:740–752.

27. Burgess S. Sample size and power calculations in Mendelian randomization with a single instrumental variable and a binary outcome. Int J Epidemiol. 2014; 43:922–929.

28. Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, et al. The MR-base platform supports systematic causal inference across the human phenome. Elife. 2018; 7:e34408.

29. Hartwig FP, Davies NM, Hemani G, Davey Smith G. Two-sample Mendelian randomization: avoiding the downsides of a powerful, widely applicable but potentially fallible technique. Int J Epidemiol. 2016; 45:1717–1726.

30. Jackson CA, Sudlow CL, Mishra GD. Education, sex and risk of stroke: a prospective cohort study in New South Wales, Australia. BMJ Open. 2018; 8:e024070.

Figure 1.
Flow diagram of the design and analysis process of this study. GWAS, genome-wide association study; AIS, any ischemic stroke; LAD, large-artery atherosclerotic disease; CE, cardiac embolism stroke; SVD, small vessel disease; MR, Mendelian randomization; SNP, single nucleotide polymorphism.
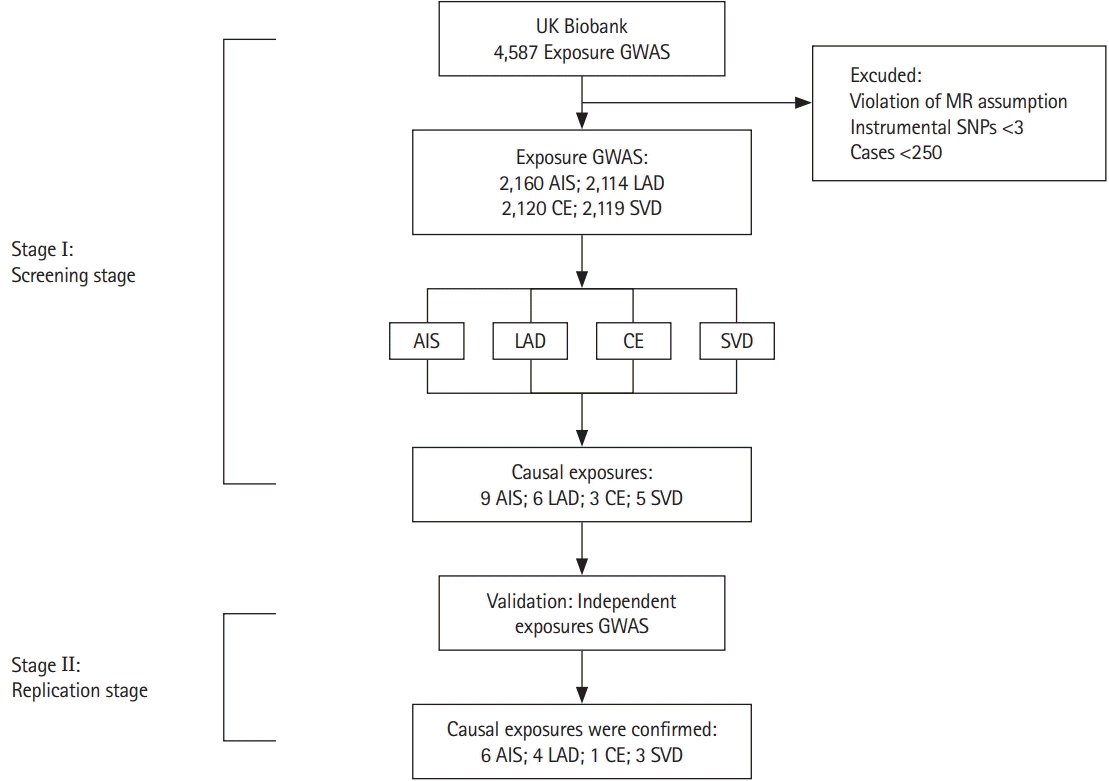
Figure 2.
Exposome-wide association studies (ExWAS) Manhattan plot highlighting exposures with statistical significance for stroke and stroke subtypes. (A) Any ischemic stroke (AIS), (B) small vessel disease (SVD), (C) large-artery atherosclerotic disease (LAD), and (D) cardiac embolism (CE). The red line indicates the exposome-wide significance threshold of the Bonferroni adjustment. The blue line indicates threshold of P<0.05.
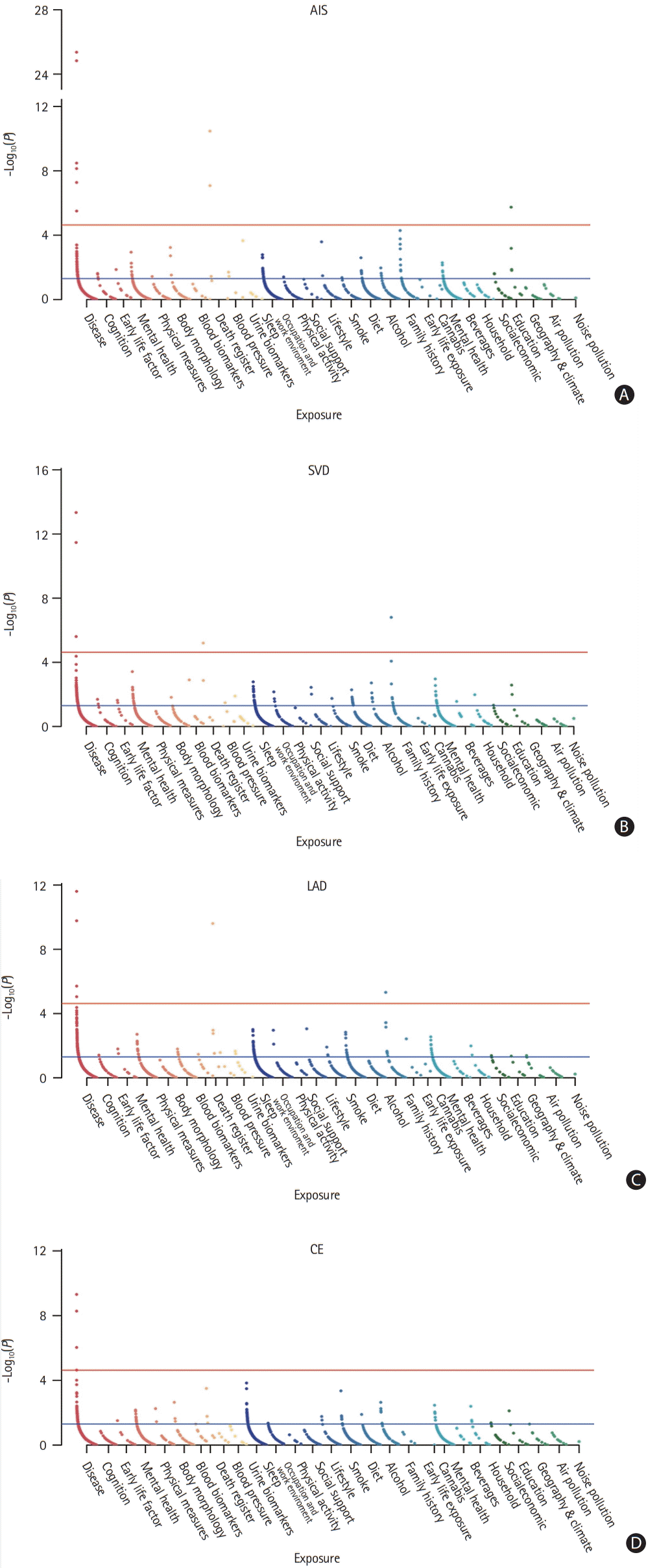
Figure 3.
Validation of the causal relationships between exposures and any stroke/stroke subtypes. The x-axis corresponds to the odds ratio (OR) for inverse-variance weighted, Mendelian randomization (MR) Egger, and weight median method. SNP, single nucleotide polymorphism; CI, confidence interval; AIS, any ischemic stroke; MTAG, multi-trait analysis of GWAS; FEV1, forced expiratory volume in 1 second; FVC, forced vital capacity; LAD, large-artery atherosclerotic disease; CE, cardiac embolism; SVD, small vessel disease.
Table 1.
Summary of significant exposures as causal mediators for stroke and stroke subtypes
SNP, single nucleotide polymorphism; GWAS, genome-wide association studies; IVW, inverse-variance weighted; AIS, any ischemic stroke; HTN, hypertension; SBP, systolic blood pressure; DM, diabetes mellitus; DBP, diastolic blood pressure; COPD, chronic obstructive pulmonary disease; LAD, large-artery atherosclerotic disease; CE, cardiac embolism; SVD, small vessel disease.
Table 2.
The causal relationships between various aspects of education and any ischemic stroke




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download