1. Ye ZW, Yuan S, Yuen KS, Fung SY, Chan CP, Jin DY. Zoonotic origins of human coronaviruses. Int J Biol Sci. 2020; 16:1686–97.

2. Pal M, Berhanu G, Desalegn C, Kandi V. Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2): an update. Cureus. 2020; 12:e7423.

3. Safiabadi Tali SH, LeBlanc JJ, Sadiq Z, Oyewunmi OD, Camargo C, Nikpour B, et al. Tools and techniques for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)/COVID-19 detection. Clin Microbiol Rev. 2021; 34:e00228–20.

4. Jolobe OMP. What are the criteria for asymptomatic status? QJM. 2021; 114:351–2.

5. LeBlanc JJ, Gubbay JB, Li Y, Needle R, Arneson SR, Marcino D, et al. Real-time PCR-based SARS-CoV-2 detection in Canadian laboratories. J Clin Virol. 2020; 128:104433.

6. Bhimraj A, Morgan RL, Shumaker AH, Lavergne V, Baden L, Cheng VC, et al. Infectious Diseases Society of America Guidelines on the treatment and management of patients with COVID-19. Clin Infect Dis. 2020 Apr 27 [Epub].
https://doi.org/10.1093/cid/ciaa478.

7. Stegeman I, Ochodo EA, Guleid F, Holtman GA, Yang B, Davenport C, et al. Routine laboratory testing to determine if a patient has COVID-19. Cochrane Database Syst Rev. 2020; 11:CD013787.

8. Nakagama Y, Komase Y, Candray K, Nakagama S, Sano F, Tsuchida T, et al. Serological testing reveals the hidden COVID-19 burden among health care workers experiencing a SARS-CoV-2 nosocomial outbreak. Microbiol Spectr. 2021; 9:e0108221.

9. Moura DTH, McCarty TR, Ribeiro IB, Funari MP, Oliveira PVAG, Miranda Neto AA, et al. Diagnostic characteristics of serological-based COVID-19 testing: a systematic review and meta-analysis. Clinics (Sao Paulo). 2020; 75:e2212.

10. Kadam SB, Sukhramani GS, Bishnoi P, Pable AA, Barvkar VT. SARS-CoV-2, the pandemic coronavirus: Molecular and structural insights. J Basic Microbiol. 2021; 61:180–202.

11. Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol. 2020; 5:562–9.

12. Xu H, Zhong L, Deng J, Peng J, Dan H, Zeng X, et al. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int J Oral Sci. 2020; 12:8.

13. Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020; 579:270–3.

14. Du L, He Y, Zhou Y, Liu S, Zheng BJ, Jiang S. The spike protein of SARS-CoV: a target for vaccine and therapeutic development. Nat Rev Microbiol. 2009; 7:226–36.

15. Saputri DS, Li S, van Eerden FJ, Rozewicki J, Xu Z, Ismanto HS, et al. Flexible, functional, and familiar: characteristics of SARS-CoV-2 spike protein evolution. Front Microbiol. 2020; 11:2112.

16. Hwang YC, Lu RM, Su SC, Chiang PY, Ko SH, Ke FY, et al. Monoclonal antibodies for COVID-19 therapy and SARS-CoV-2 detection. J Biomed Sci. 2022; 29:1.

17. Giavarina D, Carta M. Improvements and limits of anti SARS-CoV-2 antibodies assays by WHO (NIBSC 20/136) standardization. Diagnosis (Berl). 2021; 9:274–9.

18. Aguirre JJ, Ness K; Algeciras-Schimnich A. Application of the CLSI EP15-A3 guideline as an alternative troubleshooting tool for verification of assay precision. Am J Clin Pathol. 2019; 152(Suppl 1):S88.

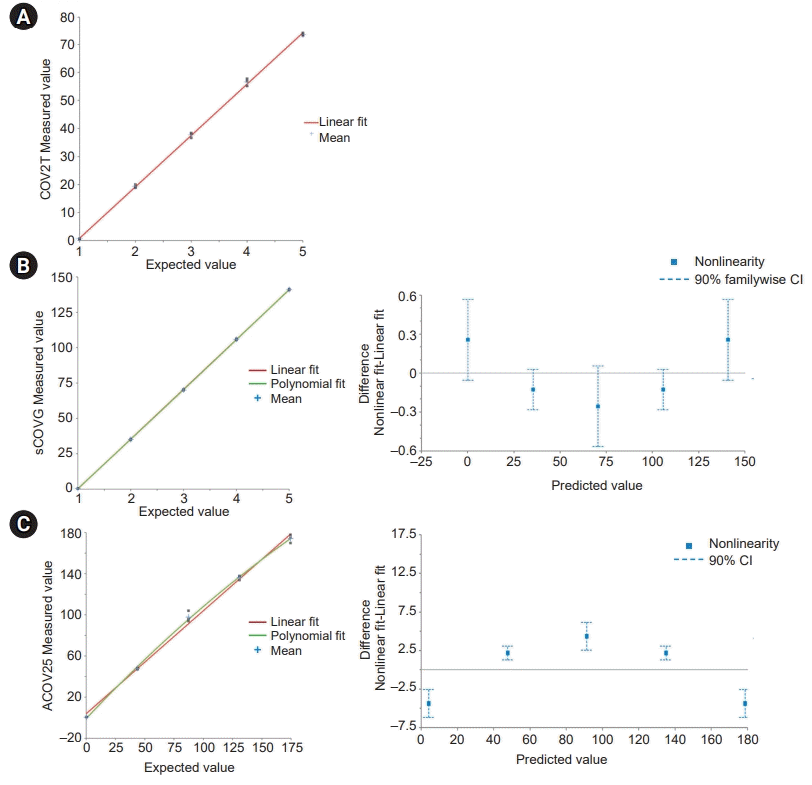
19. Tholen DW, Kroll M, Astles JR, Caffo AL, Happe TM, Krouwer J, et al. Evaluation of the linearity of quantitative measurement procedures: a statistical approach; approved guideline. Wayne: Clinical and Laboratory Standards Institute;2003.
20. Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977; 33:159–74.

21. Nittari G, Pallotta G, Amenta F, Tayebati SK. Current pharmacological treatments for SARS-COV-2: a narrative review. Eur J Pharmacol. 2020; 882:173328.

22. Trougakos IP, Stamatelopoulos K, Terpos E, Tsitsilonis OE, Aivalioti E, Paraskevis D, et al. Insights to SARS-CoV-2 life cycle, pathophysiology, and rationalized treatments that target COVID-19 clinical complications. J Biomed Sci. 2021; 28:9.

23. Tiwari N, Upadhyay J, Ansari MN, Joshi R. Novel β-coronavirus (SARS-CoV-2): current and future aspects of pharmacological treatments. Saudi Pharm J. 2020; 28:1243–52.

24. Kim MK, Lee B, Choi YY, Um J, Lee KS, Sung HK, et al. Clinical characteristics of 40 patients infected with the SARS-CoV-2 Omicron variant in Korea. J Korean Med Sci. 2022; 37:e31.

25. Leuzinger K, Osthoff M, Drager S, Pargger H, Siegemund M, Bassetti S, et al. Comparing immunoassays for SARS-CoV-2 antibody detection in patients with and without laboratory-confirmed SARS-CoV-2 infection. J Clin Microbiol. 2021; 59:e0138121.

26. Kim MA, Lee YW, Kim SR, Kim JH, Min TK, Park HS, et al. COVID-19 vaccine-associated anaphylaxis and allergic reactions: consensus statements of the KAAACI Urticaria/Angioedema/Anaphylaxis Working Group. Allergy Asthma Immunol Res. 2021; 13:526–44.

27. Igawa G, Ai T, Yamamoto T, Ito K, Nojiri S, Saito K, et al. Antibody response and seroprevalence in healthcare workers after the BNT162b2 vaccination in a University Hospital at Tokyo. Sci Rep. 2022; 12:8707.

28. Florin L, Maelegheer K, Vandewal W, Bernard D, Robbrecht J. Performance evaluation of the Siemens SARS-CoV-2 Total Antibody and IgG Antibody Test. Lab Med. 2021; 52:e147–3.

29. Haselmann V, Kittel M, Gerhards C, Thiaucourt M, Eichner R, Costina V, et al. Comparison of test performance of commercial anti-SARS-CoV-2 immunoassays in serum and plasma samples. Clin Chim Acta. 2020; 510:73–8.

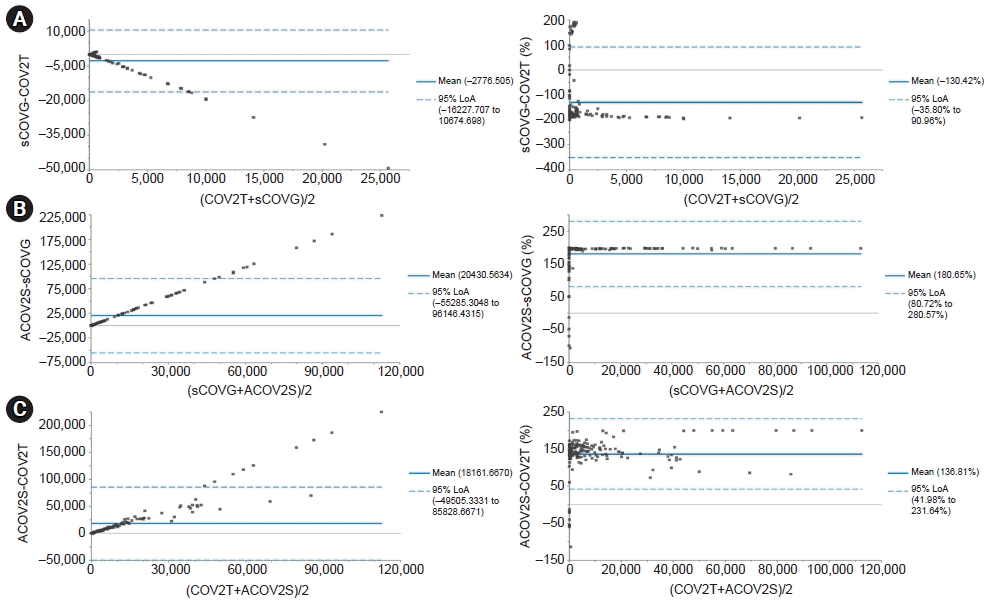
30. Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986; 1:307–10.

31. Giavarina D. Understanding Bland Altman analysis. Biochem Med (Zagreb). 2015; 25:141–51.

32. Gao J, Quan L. Current status of diagnostic testing for SARS-CoV-2 infection and future developments: a review. Med Sci Monit. 2020; 26:e928552.

33. Xu X, Sun J, Nie S, Li H, Kong Y, Liang M, et al. Seroprevalence of immunoglobulin M and G antibodies against SARS-CoV-2 in China. Nat Med. 2020; 26:1193–5.





 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download