INTRODUCTION
Glioblastoma (GBM) is the most frequent malignant brain tumor, accounting for approximately 12–15% of all intracranial neoplasms and 60–75% of astrocytic tumors. In most European and North American countries, the incidence of GBM is in the range of 3–4 new cases per 100000 populations per year
11). In Korea, an analysis of 5208 patients who were diagnosed histologically with primary central nervous system (CNS) tumors in 2010 showed that GBM accounted for 5.2% of all CNS tumors and 40.6% of all glial tumors
915). Although the overall incidence rate of primary brain tumors is very low among all human cancers, GBM is a highly invasive and aggressively growing tumor that responds poorly to radiation therapy and most forms of chemotherapy.
Temozolomide (TMZ) is an alkylating chemotherapeutic agent that causes DNA damage by transferring alkyl groups at several sites within DNA, including the O6 position of guanine. The
MGMT encodes a DNA repair protein that removes alkyl groups from O6-guanine, and thus plays a fundamental role in maintaining genomic integrity
20). Epigenetic silencing by promoter methylation results in decreased
MGMT expression and correlates with improved survival in patients with GBM who are treated with alkylating agents
4).
Although
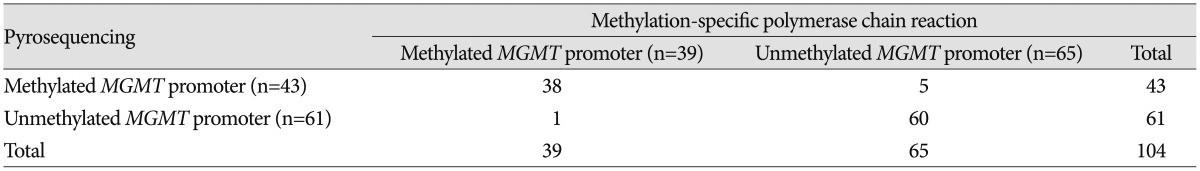
MGMT promoter methylation is used as a prognostic/predictive marker in patients with GBM, no consensus exists on the optimal analysis method. Methylation-specific polymerase chain reaction (PCR) (MSP) has been widely used, and enables cost-efficient analysis of
MGMT promoter methylation. However, it is not a quantitative method and has the risk of false-positive or false-negative results, particularly when the DNA quality and/or quantity are low. In the EORTC 26981/22981 and NCIC CE.3 trials, Hegi et al.
7) only collected data from 67% of the samples analyzed, representing 36% of cases from their archives. The success rate of MSP on formalin-fixed paraffin-embedded (FFPE) tumor samples was highly variable and the median success rate was 75.0% (range, 0% to 100%). They concluded that diagnostic MGMT testing requires sufficient and optimally preserved tumor tissue such as cryopreserved tumor specimens, thus avoiding fixation-related deterioration of tumor DNA quality. An alternative technique that overcomes the problems associated with MSP is pyrosequencing (PSQ), which is based on the sequencing-by-synthesis principle and involves a simple technique for accurate and quantitative analysis of DNA sequences. PCR followed by PSQ is a sensitive, highly reproducible, and cost-effective method for DNA methylation analyses
13). It provides absolute quantitative information based on each interrogated CpG site, which is not possible with most other methods. Additionally, unlike other methods, the assay design allows inclusion of internal controls to address inaccuracies resulting from incomplete bisulfite conversion. A group of investigators tried to compare and optimize three quantitative
MGMT promoter methylation techniques and concluded that the PSQ assay is the most accurate and is also robust when applied to archival samples, including those of GBM
14).
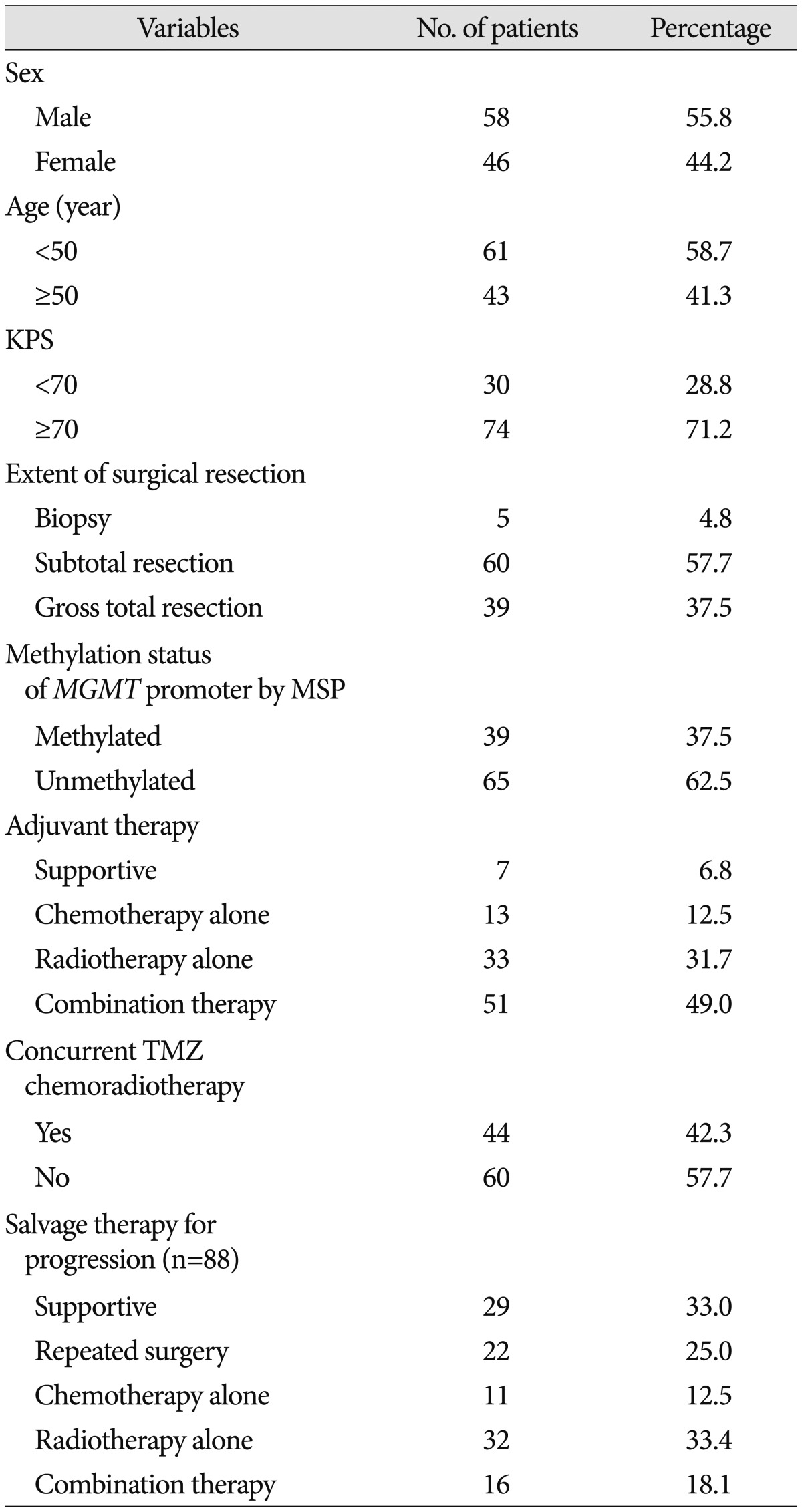
The objective of this study was to determine whether PSQ could be used to determine quantitatively the methylation status of the MGMT promoter as a clinical biomarker in routine practice using 2- to 15-year-old archival tissue samples of GBMs. We also estimated the methylation status of the MGMT promoter using MSP, which is already known to be a prognostic marker for overall survival and a predictive marker for conventional treatment of GBM patients, and verified the results obtained with PSQ. In addition, we examined other prognostic factors for survival of GBM patients.
Go to :

DISCUSSION
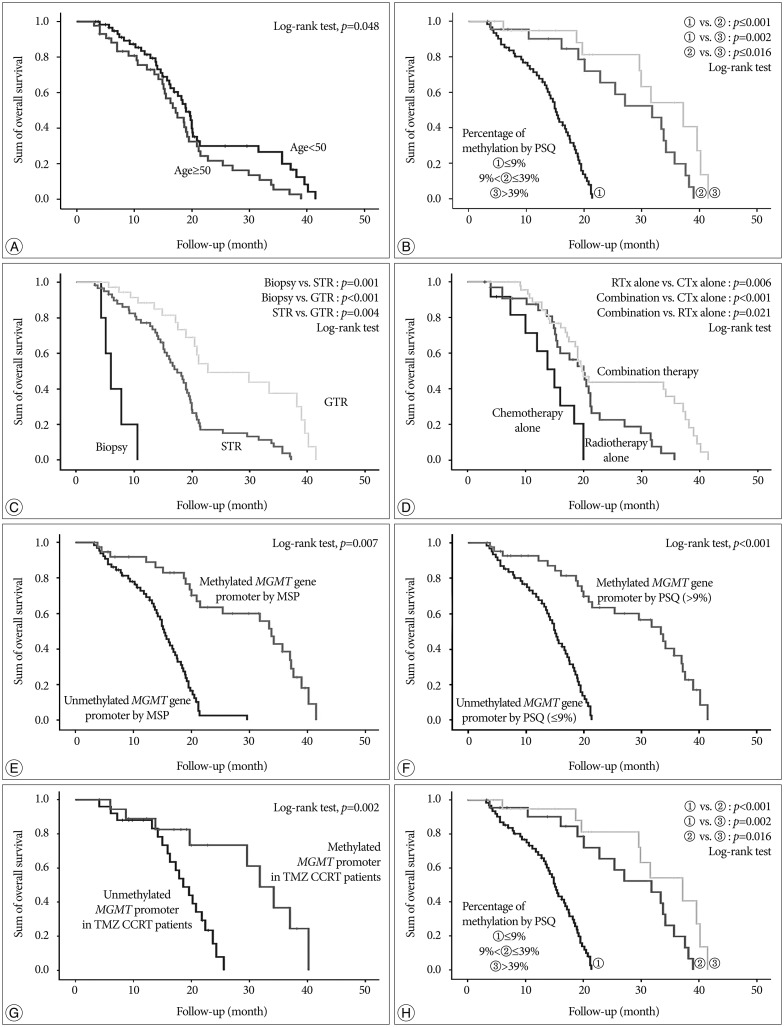
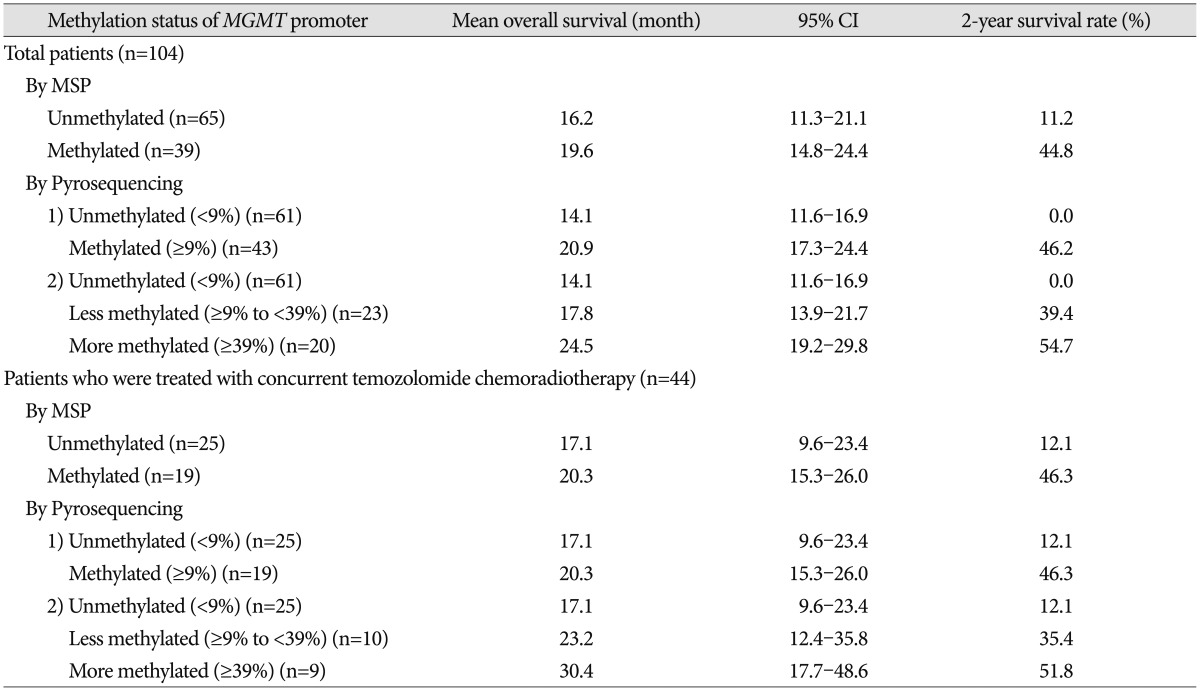
In this study, we performed PSQ analysis to determine MGMT promoter methylation status and estimate the extent of methylation using 1- to 13-year-old archival tissue samples of GBMs. We also investigated the prognostic significance of MGMT promoter methylation and other factors associated with OS in patients who were treated in the routine clinic.
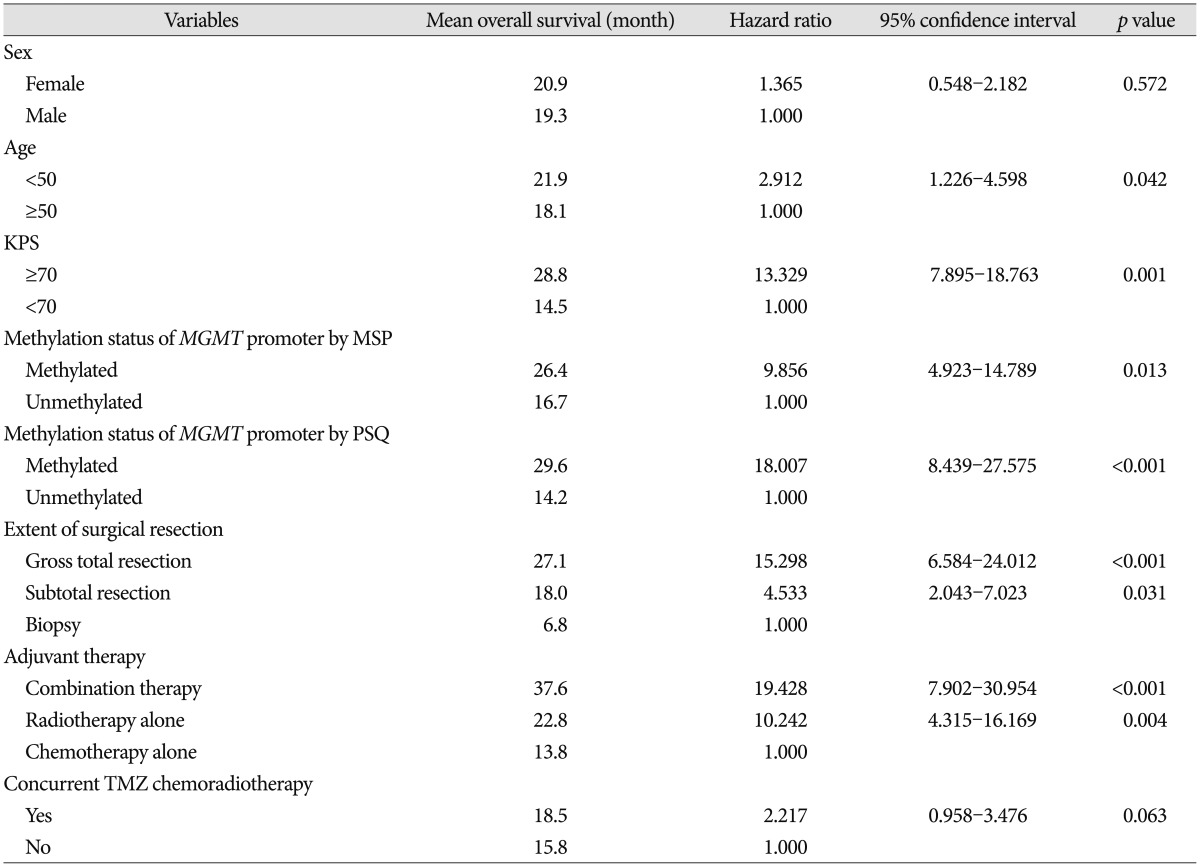
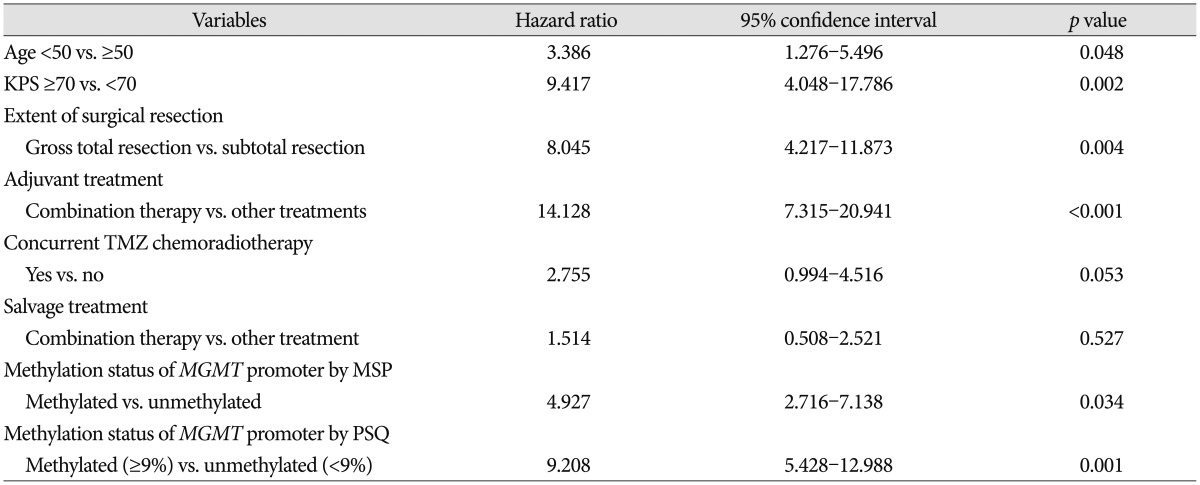
Recently, Stupp et al.
19) reported the results of 5-year analysis of the EORTC-NCIC trial for survival in GBM patients. Their randomized phase III study showed that the following factors had an association with outcome : concurrent chemoradiotherapy with TMZ, complete resection, age <50, good functional status (recursive partitioning analysis class III>IV>V), and methylated
MGMT promoter. We obtained similar results in our study; age <50, good functional status (KPS ≥70), gross total resection, and methylated
MGMT promoter analyzed by both MSP and PSQ were all associated with OS. Therefore, this study provided good verification of the published literature.
In terms of the frequency of
MGMT promoter methylation in GBM, Hegi et al.
7) reported that 45% of GBMs showed
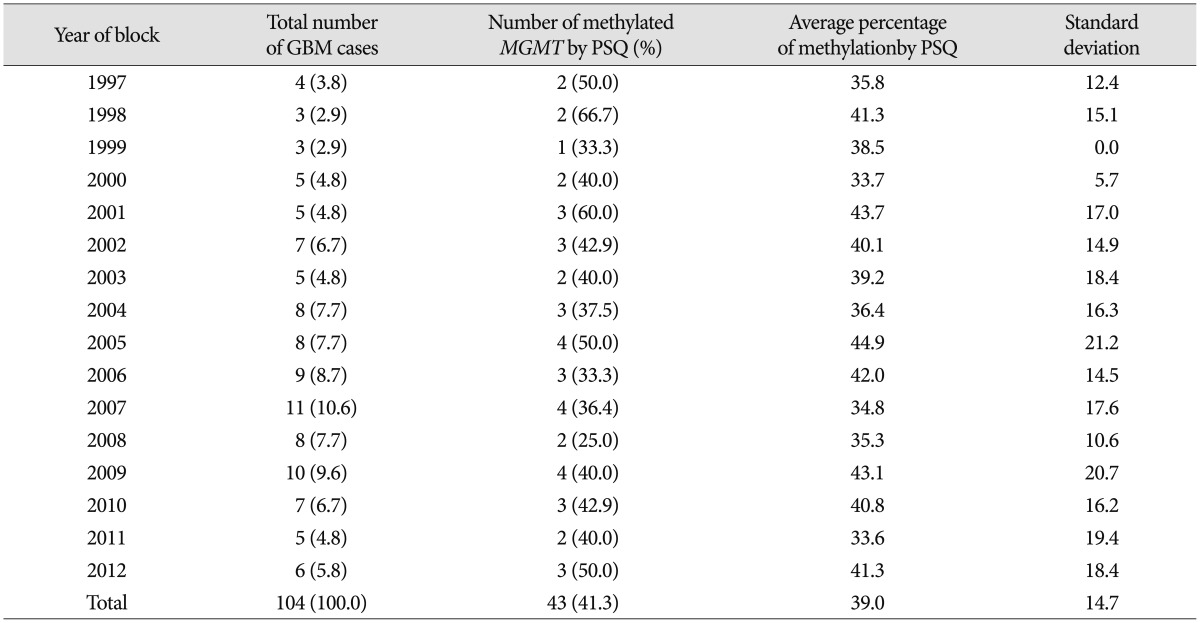
MGMT promoter methylation in their randomized trial, and that the median success rate of MSP analysis in FFPE samples was 75%. In contrast, the success rate of PSQ analysis in our study was 100% for 104 samples from 1- to 13-year-old archival specimens, and 41.3% of the GBMs were methylated. Recently, Lee et al.
12) reported that 44.4% of 27 GBMs showed methylation. These results, and data from several other researchers, indicate that 40–50% of GBMs are potentially methylated, which might be of clinical benefit regarding treatment with alkylating agents.
Several methods for determining promoter methylation have been described, including the use of restriction enzymes
18) or genomic bisulfite sequencing
6). Among these methods, some are quantitative, some are semi-quantitative, and some are qualitative. The overwhelming majority of published data uses MSP following bisulfite treatment
8). The MSP assay is highly sensitive and has been used widely in this context
18). However, as there are no inbuilt measures of the adequacy of bisulfite treatment, the possibility of false positives due to inadequate conversion of non-methylated cytosine to uracil exists, particularly for DNA of poor quality or low quantity. Another potential source of false positives is mis-priming, which may be a greater problem when high numbers of PCR cycles or nested primers are used. An accurate quantitative approach is very important for DNA methylation analysis, and MSP has many weaknesses in this regard. One of the major concerns with MSP is the use of relative quantification by means of an external control using a different PCR for the control gene. The accuracy and reproducibility of this approach for the large diversity of genes has still not been adequately addressed. Development of DNA methylation risk prediction panels based on diverse genes will be required to develop and optimize an accurate and reproducible method.
In this study, we present results of PSQ analysis of 1- to 13-year-old archive specimens. As described above, PSQ is a simple technique based on the sequencing-by-synthesis principle and allows accurate and quantitative analysis of DNA sequences. The percentage of methylation was evenly distributed over the years of this study and no increasing or decreasing pattern of methylation with specimen age was observed, regardless of the age of the samples. This might represent a huge improvement in the success rate when compared with data reported using MSP by Hegi et al.
7). Using the same method as in the current study, Dunn et al.
2) analyzed 264 of 287 tumor samples from 121 cases by PSQ. Some failures occurred, most likely as a result of the small sample volume (<1 mm
3) with low DNA yield or poor DNA integrity in the FFPE samples. Nonetheless, despite these failures the PSQ data were reproducible and showed a good correlation between duplicate PCR reactions from the same bisulfite modification and between two independent bisulfite modifications of the same DNA extract
2). More recently, a quantitative real-time MSP assay in which the copy number of methylated
MGMT alleles was calculated showed improvement over the gel-based MSP assay in terms of reproducibility and use with archival samples, but did not provide methylation data at individual CpG sites
21). Therefore, PCR followed by the PSQ method is a stable technique that produces consistent data. These results support the need for a large nationwide retrospective trial to evaluate the
MGMT promoter methylation status of old archived material from multiple institutions.
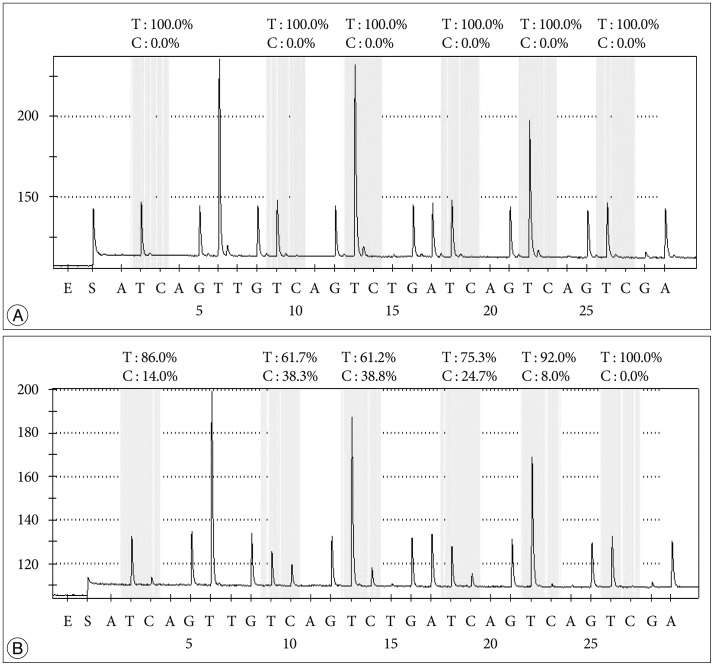
In terms of the relationship between the extent of methylation estimated by PSQ and clinical outcome in GBM, this study showed that GBM patients with a more methylated
MGMT promoter had longer overall survival than those with a less methylated
MGMT promoter. Similarly, Dunn et al.
2) reported an interesting clustering analysis based on the extent of methylation. Progression-free survival was significantly different between low (mean percentage of methylation 9–29%) and high (≥29%) methylation groups of GBM, and between high methylated and unmethylated cases. Log-rank tests showed significant differences in OS between unmethylated and methylated groups and between cases with a low versus high percentage of methylation. This result suggested that qualitative and quantitative data should be simultaneously obtained from the
MGMT promoter methylation analysis to predict progression-free survival, overall survival, or chemoresponsiveness in clinical practice. Although we did not obtain exactly the same results as those of Dunn, we found a similar tendency for a higher percentage of methylation to be associated with longer overall survival. The discrepancy in results probably originates from different sample size and different follow-up duration. Moreover, our archives included a relatively small number of samples with a low percentage of methylation.
Another issue for
MGMT promoter methylation analysis is the relevant CpG sites for methylation status. The pattern of
MGMT promoter methylation is very heterogeneous from tumor to tumor, and the specific CpG sites that need to be methylated to silence transcription and provide a favorable outcome have not been established. Several studies have investigated the methylation status at individual CpGs within
MGMT CGIs and compared them with gene expression. Watts et al.
22) studied the methylation status of 108 CpGs across
MGMT CGIs using bisulfite sequencing and identified three distinct regions within the CGI where high methylation levels were found, but only in the
MGMT non-expressing cell line. Recently, Karayan-Tapon et al.
10) assessed
MGMT promoter methylation status by five different methods including PSQ, and the PSQ primer sets used included 5 CpGs in the DMR2 region that were also used in this study. They concluded that PSQ using FFPE samples predicted overall survival well in patients who were initially treated with radiotherapy plus TMZ.
The limitations of this study can be summarized as follows : First, the cut-off value for methylation used in this study should be verified in more detail and in a larger number of samples. Second, we studied limited sites of CGI methylation using the Pyro Mark Q96 CpG MGMT kit (which evaluates only 5 CGIs). Therefore, further evaluation of other sites of CGI methylation is essential to increase the accuracy of the status and extent of MGMT promoter methylation, and to commercialize this kit. Third, the relatively small number of GBM samples made it impossible for us to find any critical methylation status value for predicting the outcome of these tumors. Fourth, this study is neither a randomized study nor a prospective study. Finally, progression-free survival could not be estimated because there was no specific protocol for follow-up and management.
Go to :





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download