Abstract
Moyamoya disease (MMD) is an arteriopathy of the intracranial circulation predominantly affecting the branches of the internal carotid arteries. Heterogeneity in presentation, progression and response to therapy has prompted intense study to improve the diagnosis and prognosis of this disease. Recent progress in the development of moyamoya-related biomarkers has stimulated marked interest in this field. Biomarkers can be defined as biologically derived agents-such as specific molecules or unique patterns on imaging-that can identify the presence of disease or help to predict its course. This article reviews the current categories of biomarkers relevant to MMD-including proteins, cells and genes-along with potential limitations and applications for their use.
Moyamoya is a progressive arteriopathy of unclear etiology that predominantly affects the major branches of the internal carotid arteries5868). First described in Japan, this disorder was initially characterized by radiographic criteria, as defined by Suzuki and Takaku68). These elegant anatomic studies have served as the foundation underlying decades of clinical and laboratory efforts to better understand this disease. Over time, it has increasingly become apparent that these angiographic findings represent a multitude of distinct pathophysiologic processes that manifest a shared radiographic signature958).
Initially, this concept of multiple conditions culminating in a common end pathway was acknowledged by the distinction between moyamoya "disease" (bilateral arteriopathy existing in isolation) and moyamoya "syndrome" (either unilateral arteriopathy or arteriopathy found in conjunction with some other medical disorder)958). By convention, the arteriopathies are usually collectively referred to as "moyamoya disease" (MMD). This awareness of heterogeneity in clinical presentation has prompted research focused on the development of techniques capable of better defining MMD. One area with the potential to advance the diagnosis, prognosis and understanding of MMD is the field of biomarkers.
Biomarkers are defined as "any substance, structure, or process that can be measured in the body or its products and influence or predict the incidence of outcome or disease"66). In essence, biomarkers are biological "fingerprints" that can be used by clinicians to identify a specific disease. Broadly speaking, this definition can include physiological signs that are associated with a clinical phenotype, anatomic structures that can be identified radiographically as a distinct signature, molecular "fingerprints" of specific proteins measured in patient samples, unique cell types associated with disease or genetic mutations. This manuscript summarizes current biomarkers relevant to the diagnosis of MMD, including phenotypes, radiographic findings, proteins, circulating cells and genetic/epigenetic markers, followed by discussion of how they could be employed in practice.
Moyamoya disease was first described in 1957 in Japan when the clinical symptoms of cerebral infarction were found in conjunction with "hypoplasia" of the internal carotid arteries70). Over the following 12 years, increasing recognition of common patient presentations allowed clinicians to couple specific symptoms with a distinct radiographic pattern, as ultimately reported by Suzuki68). While the need to rely solely on clinical observation to define phenotypes is being supplanted by imaging, molecular profiling and genetic analysis, awareness of associations between specific medical conditions and MMD is still important in practice.
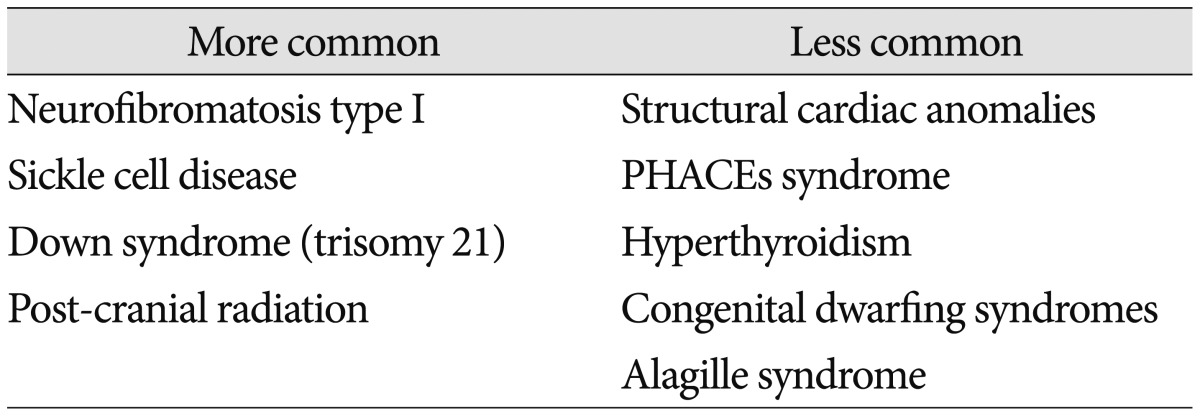
Although only loosely contained within the rubric of biomarkers, a list of medical conditions linked to MMD is useful. In addition to the practical application of helping physicians recognizing phenotypes that may be at-risk for arteriopathy, translational research efforts can be informed by clinical observation. Table 1 outlines some of the more common disorders that have been reported in conjunction with MMD (Table 1)362934354751586171).
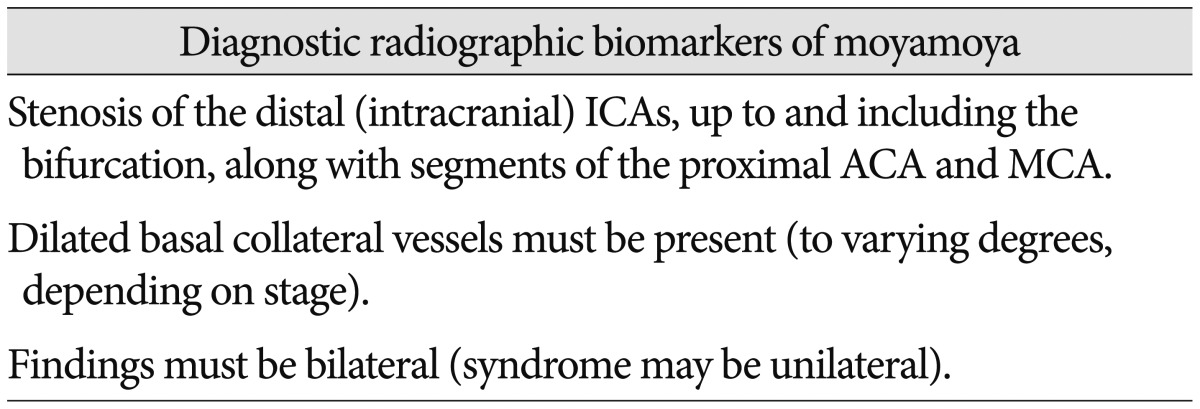
Radiographic biomarkers have emerged as critical tools to identify disease and stratify risk in patients64). The initial description of MMD is predicated on a radiographic signature, the "puff of smoke" on catheter arteriography68). Since then, this radiographic biomarker has been used as the primary foundation upon which to build a definition of MMD963). Specific characteristics have been codified in order to assist clinical decision making, such as determining risk of stroke, chance of hemorrhage, likelihood of co-existing genetic conditions and need for surgery91114252630). There is great variability in the current utilization of radiographic biomarkers across institutions that care for patients with MMD. However, a number of key markers have been cited repeatedly and have gained general acceptance. These signature findings are summarized in Table 2 and have been adopted in the International Classification of Diseases (Table 2)95863).
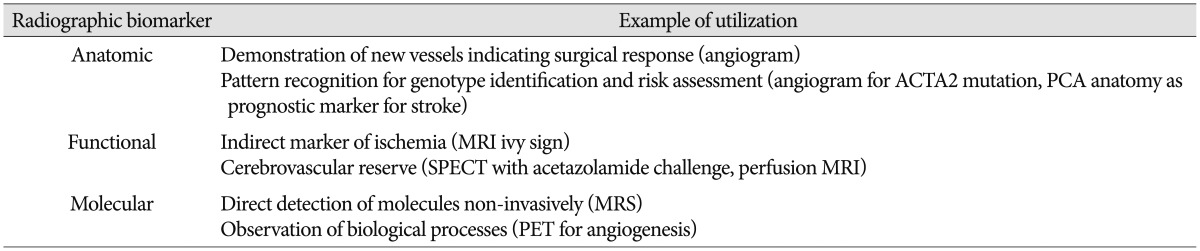
Radiographic biomarkers for MMD can be utilized in a wide array of clinical scenarios (Table 3). Unique patterns of vessel branching on arteriogram may indicate specific mutations, such as ACTA2 (discussed below)48). Distinct anomalies, such as posterior cerebral artery stenosis, may portend a higher risk of hemorrhage14). Any vessel anomaly identified by angiography on the non-affected hemisphere in unilateral MMD may increase the chance of subsequent progression to bilateral disease2862). Changes in radiographic studies may predict response to therapy more accurately and can be used in formulating follow-up plans for patients10253763). In addition to functional perfusion studies, indirect measures of blood flow, such as reduction in the "ivy sign" on MRI may serve to mark a successful response to surgery827). Specific molecules can be detected with magnetic resonance spectroscopy and these metabolites may aid in prognosis13). More recently, imaging of patients pre- and post-operatively using positron emission tomography (PET) modified with novel labeled peptides has improved the potential to non-invasively observe the biological process of angiogenesis533). These findings underscore the immense potential of radiographic biomarkers to aid in the care of patients with MMD.
Measuring levels of proteins in body tissues or fluids as biomarkers requires that the proteins are somehow related to the presence of the disease, either directly or indirectly. It can sometimes be difficult to ascertain if the putative biomarker is produced directly from the pathologic tissue (such as prolactin from a prolactinoma) or as a secondary response of the body to the disease (such as C-reactive protein in the setting of a bacterial infection). Where one looks for a biomarker-tissue, spinal fluid, serum, urine, saliva-is usually secondary to discovering a clinically relevant molecule. Generally speaking, there are two major approaches to biomarker discovery; hypothesis driven discovery (in which specific molecules are selected a priori due to suspected roles in the given pathophysiologic processes) and proteomic screening (in which the entire proteome of specimens from diseased patients are compared to matched controls to reveal differences in expression).
In looking for candidate biomarkers for MMD, both approaches have been applied. Initial efforts focused largely on the cerebrospinal fluid (CSF), given the proximity to the disease process in the cerebral vessels. General proteome analysis of the CSF has been applied by many groups around the world24356656975). These studies have identified a number of expected candidate molecules related to ischemia and angiogenesis (as outlined below), but also some molecules of unclear pathophysiologic relevance, including oxyntomodulin, urocortin-2, beta-defensin 133, antibacterial protein LL-37 and liver-expressed antimicrobial peptide 22). One of the most interesting recent applications of mass spectrometry in MMD biomarker discovery was the report of a novel peptide, 4473Da, that appeared to correlate with favorable postoperative collateral development in a small cohort of patients43). The significance of these findings continues to be assessed in studies with larger cohorts of patients.
Another method of organizing biomarker discovery is to categorize subgroups of molecules by function. Proteins linked to MMD include the broad classes of enzymes, growth factors, adhesion molecules and inflammatory/coagulation peptides (summarized in Table 4). These often result from hypothesis-driven biomarker discovery, but it can be difficult to ascertain if the molecules are uniquely causative of the disease or secondary by-products of the arteriopathy. While these markers have utility within the context of MMD, it will be important to direct future research toward determining which markers (if any) comprise a MMD-specific "fingerprint" and which might be more non-specific markers of general physiological processes like ischemia or angiogenesis.
Circulating cells have been employed as markers of disease in other fields, most notably in cancer research. Recently, these efforts have been expanded to include MMD. Central to the premise of this work is the hypothesis that MMD involves ongoing vascular remodeling, including both the primary arteriopathy and also the secondary angiogenesis from collateral development. Consequently, investigators have searched for endothelial cells and smooth muscle cells involved in these processes.
Pathological studies have long shown that proliferation of smooth muscle cells in the walls of the affected arteries in MMD is a common finding44). Smooth muscle progenitor cells (SPCs) have been isolated from the blood of patients with MMD. Recent analysis of these cells show that SPCs from moyamoya patients demonstrate irregular tube formation in assays when compared to SPCs from matched controls24). In addition, these SPCs exhibited differential expression of over 200 genes, including reduced CD31 expression, relative to controls from healthy individuals24). The ability to isolate a specific cell type and demonstrate differences in both protein expression and cell function suggest a dynamic approach to biomarker discovery in MMD.
Parallel to this work with smooth muscle cells, investigation into the role of endothelial cells as markers of MMD has also yielded some provocative results. Migration of these cells into the intima of the internal carotid at the site of stenosis in MMD has been suggested by pathological studies and it is hypothesized that these cells may play a role in both proximal arterial narrowing as well as distal collateral development67). CD34+ cells, a subpopulation of endothelial progenitor cells, have been reported to be detected at increased levels in the blood of patients with MMD when compared to healthy controls and also when compared to patients with non-MMD intracranial arterial stenosis5074). However, conflicting data has been reported when specifically looking at CD34+ cells in children, in which another group has reported decreased levels of CD34+ cells in MMD patients relative to matched controls31). Adding to the complexity of biomarker development, research has also been undertaken assessing not just the quantity of cells present, but also evaluating their function, as measured by assays of tube formation and colony formation2031). It has been proposed that these circulating endothelial cells exhibit reduced function when assessed in vitro relative to those from healthy controls.
The use of living cells as biomarkers for MMD is inherently more complicated than measurement of protein levels or genetic analysis. Variability in isolation of specific subpopulations of cells, the dynamic nature of cell marker expression and the lack of standardization in measures of cell function are technical issues that currently limit the utility of this approach. However, the concept of cell-based biomarkers holds tremendous appeal and it is expected that future efforts will mitigate many of these problems. Ultimately, circulating cells may provide the best combination of proteomic, genomic and functional markers of disease.
The ultimate goal of biomarkers is to unequivocally identify the presence or absence of disease with a high degree of sensitivity and specificity. Discovery of a genetic mutation that is reproducibly linked with a distinct disease phenotype is one the most sought-after objectives in biomedical research. The ability to associate specific genes with MMD is complicated by the likely heterogeneity of disorders that share a common phenotype58). Initial efforts to discover genetic markers of MMD reflected this complexity, with a wide range of chromosomes, genes and hereditary diseases reported to be putative markers (Table 5).
While these early attempts may have met with mixed success- and may yet yield important discoveries for individual subtypes of MMD syndrome-two major advances in the genetics of MMD recently been reported and validated by several groups. First, the discovery of R179 mutations in ACTA2 revealed that specific mutations in genes specific to smooth muscle cells of the vasculature can reliably manifest an arteriopathy identifiable as moyamoya1148). However, it rapidly became apparent that this mutation was associated with only a small minority of MMD cases, as evidenced by studies in Asia and Europe5560). The second major genetic biomarker of MMD was a mutation in the gene for RNF213224046). While the function of the protein encoded by the gene remains to be confirmed (potentially a regulator of an ATPase in smooth muscle cells), population studies suggest that this is a major contributor to MMD disease in patients of Asian ancestry, present in up to 90% of familial cases in Asia. Moreover, specific mutations in this gene may also help to improve prognosis, with one base-pair mutation predicting a severe, early-onset form of moyamoya46).
These new biomarkers are changing the practice of physicians who treat patients with MMD, informing clinical decisions and helping to predict familial risk. While guidelines remain in evolution, the impact of this sort of biomarker discovery is clear. Future efforts will need to refine subgroups of MMD by genetic mutational analysis, define function of relevant genes and evaluate potential epigenetic factors that may serve as important modulators of disease phenotype5253).
Disease-specific biomarkers are only important if they confer benefits to patients in clinical practice. "The key issue at hand is determining the relationship between any given measurable biomarker and relevant clinical endpoints"66). Ideally, biomarkers should aid in the diagnosis, prognosis or treatment of disease. Consequently, discussion of the potential use of MMD-specific biomarkers is a critical factor guiding their development and implementation.
Accurate and timely diagnosis of MMD is critically important. The single factor that overwhelmingly influences long-term outcome is the neurological status at time of treatment59). Data from patients with early diagnosis of MMD prior to devastating stroke supports the premise that the ability to make an early diagnosis of the arteriopathy would profoundly improve the outcome of patients3540). As described here, there are several biomarker-related approaches that can directly impact this objective.
Refinement of clinical phenotypes that predict at-risk populations is ongoing and the first step in selecting individuals as candidates for further testing. Imaging remains the gold standard for confirming the diagnosis of MMD, but widespread utilization of imaging studies is cost-prohibitive. In contrast, the development of cheaper, non-invasive screening tests predicated on biomarkers able to detect MMD would revolutionize the care of affected patients. Measurement of proteins in serum, blood or urine would be particularly useful for this goal, as would genetic testing. ELISA and gene sequencing technologies are readily available, relatively low-cost and could markedly complement the use of imaging studies.
The prognosis of MMD continues to challenge physicians. Some populations are prone to rapid, fulminant decline (such as very young infants), while other groups may manifest a far more indolent course19363772). Development of biomarkers to better predict those patients that would benefit from surgery is an area of interest. Radiographic biomarkers may be particularly useful for this objective. For example, data enabling surgeons to prospectively identify higher risk of contralateral progression (in unilateral MMD) or increased likelihood of posterior circulation stroke is already influencing practice patterns394062).
Biomarkers may also impact care by improving therapeutic efficacy. Studies of cell function may help to predict the capacity for therapeutic angiogenesis, and these data could be used to inform the decision about choosing direct or indirect bypass. Measures of circulating peptides may provide additional data to more accurately predict response to surgery. Ultimately, changes in measured biomarker levels may suggest novel therapeutic agents, such as growth factors that could be used to modulate surgical collateral growth2338).
Moving forward, biomarker development for MMD has tremendous potential, but also faces challenges. The relative rarity of the disease means that collaboration between centers will be important in validating candidate biomarkers. Shared data, longitudinal studies and comparative trials will be crucial to generating meaningful results. Equally important will be efforts to generate consensus on how to best utilize the new data provided by biomarkers. Clear articulation of the strengths and weaknesses of new diagnostic and prognostic capabilities afforded by research will help to avoid unrealistic expectations. Laboratory efforts should be complemented by regular meetings of working groups focused on objectively reviewing progress on biomarker applications in the clinic.
MMD-specific biomarkers include clinical phenotypes, radiographic signatures, patterns of protein expression, distinct circulating cell populations and specific genetic mutations. Generally, evidence of characteristic narrowing of the anterior cerebral circulation, reduction in cortical perfusion, elevations in angiogenesis-related peptides and alterations in circulating endothelial cell function are common findings. Several genetic associations have been described, with two recently reported specific mutations (in ACTA2 and RNF213) that manifest distinct clinical presentations. It is increasingly apparent that the term "moyamoya" encompasses many different pathophysiologic conditions and that the use of biomarkers will refine and improve our understanding of this arteriopathy. Future efforts will benefit from multicenter studies and working groups to help guide adoption of utilization in clinical practice.
Acknowledgements
Chae Moyamoya Research Fund, Kids at Heart Neurosurgery Research Foundation, Justin Doo Research Fund.
References
1. Amano T, Inoha S, Wu CM, Matsushima T, Ikezaki K. Serum alpha1-antitrypsin level and phenotype associated with familial moyamoya disease. Childs Nerv Syst. 2003; 19:655–658. PMID: 12955420.

2. Araki Y, Yoshikawa K, Okamoto S, Sumitomo M, Maruwaka M, Wakabayashi T. Identification of novel biomarker candidates by proteomic analysis of cerebrospinal fluid from patients with moyamoya disease using SELDI-TOF-MS. BMC Neurol. 2010; 10:112. PMID: 21059247.

3. Baird LC, Smith ER, Ichord R, Piccoli DA, Bernard TJ, Spinner NB, et al. Moyamoya syndrome associated with Alagille syndrome : outcome after surgical revascularization. J Pediatr. 2015; 166:470–473. PMID: 25465847.

4. Bernard TJ, Fenton LZ, Apkon SD, Boada R, Wilkening GN, Wilkinson CC, et al. Biomarkers of hypercoagulability and inflammation in childhood-onset arterial ischemic stroke. J Pediatr. 2010; 156:651–656. PMID: 20022340.

5. Choi H, Phi JH, Paeng JC, Kim SK, Lee YS, Jeong JM, et al. Imaging of integrin α(V)β(3) expression using (68)Ga-RGD positron emission tomography in pediatric cerebral infarct. Mol Imaging. 2013; 12:213–217. PMID: 23651498.
6. Codd PJ, Scott RM, Smith ER. Seckel syndrome and moyamoya. J Neurosurg Pediatr. 2009; 3:320–324. PMID: 19338412.

7. Fujimura M, Watanabe M, Narisawa A, Shimizu H, Tominaga T. Increased expression of serum Matrix Metalloproteinase-9 in patients with moyamoya disease. Surg Neurol. 2009; 72:476–480. discussion 480PMID: 19147196.

8. Fujiwara H, Momoshima S, Kuribayashi S. Leptomeningeal high signal intensity (ivy sign) on fluid-attenuated inversion-recovery (FLAIR) MR images in moyamoya disease. Eur J Radiol. 2005; 55:224–230. PMID: 16036151.

9. Fukui M. Guidelines for the diagnosis and treatment of spontaneous occlusion of the circle of Willis ('moyamoya' disease). Research Committee on Spontaneous Occlusion of the Circle of Willis (Moyamoya Disease) of the Ministry of Health and Welfare, Japan. Clin Neurol Neurosurg. 1997; 99(Suppl 2):S238–S240. PMID: 9409446.
10. Goda M, Isono M, Ishii K, Kamida T, Abe T, Kobayashi H. Long-term effects of indirect bypass surgery on collateral vessel formation in pediatric moyamoya disease. J Neurosurg. 2004; 100(2 Suppl Pediatrics):156–162. PMID: 14758943.

11. Guo DC, Papke CL, Tran-Fadulu V, Regalado ES, Avidan N, Johnson RJ, et al. Mutations in smooth muscle alpha-actin (ACTA2) cause coronary artery disease, stroke, and Moyamoya disease, along with thoracic aortic disease. Am J Hum Genet. 2009; 84:617–627. PMID: 19409525.

12. Han H, Pyo CW, Yoo DS, Huh PW, Cho KS, Kim DS. Associations of Moyamoya patients with HLA class I and class II alleles in the Korean population. J Korean Med Sci. 2003; 18:876–880. PMID: 14676447.

13. Harada M, Miyoshi H, Uno M, Okada T, Hisaoka S, Hori A, et al. Neuronal impairment of adult moyamoya disease detected by quantified proton MRS and comparison with cerebral perfusion by SPECT with tc-99m HM-PAO : a trial of clinical quantification of metabolites. J Magn Reson Imaging. 1999; 10:124–129. PMID: 10441014.

14. Hishikawa T, Tokunaga K, Sugiu K, Date I. Clinical and radiographic features of moyamoya disease in patients with both cerebral ischaemia and haemorrhage. Br J Neurosurg. 2013; 27:198–201. PMID: 22934580.

15. Hojo M, Hoshimaru M, Miyamoto S, Taki W, Nagata I, Asahi M, et al. Role of transforming growth factor-beta1 in the pathogenesis of moyamoya disease. J Neurosurg. 1998; 89:623–629. PMID: 9761057.

16. Hong SH, Wang KC, Kim SK, Cho BK, Park MH. Association of HLA-DR and -DQ genes with familial moyamoya disease in Koreans. J Korean Neurosurg Soc. 2009; 46:558–563. PMID: 20062572.

17. Ikeda H, Sasaki T, Yoshimoto T, Fukui M, Arinami T. Mapping of a familial moyamoya disease gene to chromosome 3p24.2-p26. Am J Hum Genet. 1999; 64:533–537. PMID: 9973290.

18. Inoue TK, Ikezaki K, Sasazuki T, Matsushima T, Fukui M. Linkage analysis of moyamoya disease on chromosome 6. J Child Neurol. 2000; 15:179–182. PMID: 10757474.

19. Jackson EM, Lin N, Manjila S, Scott RM, Smith ER. Pial synangiosis in patients with moyamoya younger than 2 years of age. J Neurosurg Pediatr. 2014; 13:420–425. PMID: 24527861.

20. Jung KH, Chu K, Lee ST, Park HK, Kim DH, Kim JH, et al. Circulating endothelial progenitor cells as a pathogenetic marker of moyamoya disease. J Cereb Blood Flow Metab. 2008; 28:1795–1803. PMID: 18612318.

21. Kainth DS, Chaudhry SA, Kainth HS, Suri FK, Qureshi AI. Prevalence and characteristics of concurrent down syndrome in patients with moyamoya disease. Neurosurgery. 2013; 72:210–215. discussion 215PMID: 23149966.

22. Kamada F, Aoki Y, Narisawa A, Abe Y, Komatsuzaki S, Kikuchi A, et al. A genome-wide association study identifies RNF213 as the first Moyamoya disease gene. J Hum Genet. 2011; 56:34–40. PMID: 21048783.

23. Kang HS, Kim JH, Phi JH, Kim YY, Kim JE, Wang KC, et al. Plasma matrix metalloproteinases, cytokines and angiogenic factors in moyamoya disease. J Neurol Neurosurg Psychiatry. 2010; 81:673–678. PMID: 19965844.

24. Kang HS, Moon YJ, Kim YY, Park WY, Park AK, Wang KC, et al. Smooth-muscle progenitor cells isolated from patients with moyamoya disease : novel experimental cell model. J Neurosurg. 2014; 120:415–425. PMID: 24160477.

25. Kashiwagi S, Yamashita T, Katoh S, Kitahara T, Nakashima K, Yasuhara S, et al. Regression of moyamoya vessels and hemodynamic changes after successful revascularization in childhood moyamoya disease. Acta Neurol Scand Suppl. 1996; 166:85–88. PMID: 8686450.
26. Kawaguchi S, Sakaki T, Morimoto T, Kakizaki T, Kamada K. Characteristics of intracranial aneurysms associated with moyamoya disease. A review of 111 cases. Acta Neurochir (Wien). 1996; 138:1287–1294. PMID: 8980731.

27. Kawashima M, Noguchi T, Takase Y, Nakahara Y, Matsushima T. Decrease in leptomeningeal ivy sign on fluid-attenuated inversion recovery images after cerebral revascularization in patients with Moyamoya disease. AJNR Am J Neuroradiol. 2010; 31:1713–1718. PMID: 20466798.

28. Kelly ME, Bell-Stephens TE, Marks MP, Do HM, Steinberg GK. Progression of unilateral moyamoya disease : a clinical series. Cerebrovasc Dis. 2006; 22:109–115. PMID: 16685122.

29. Kennedy BC, McDowell MM, Yang PH, Wilson CM, Li S, Hankinson TC, et al. Pial synangiosis for moyamoya syndrome in children with sickle cell anemia : a comprehensive review of reported cases. Neurosurg Focus. 2014; 36:E12. PMID: 24380478.
30. Khan N, Yonekawa Y. Moyamoya angiopathy in Europe. Acta Neurochir Suppl. 2005; 94:149–152. PMID: 16060256.

31. Kim JH, Jung JH, Phi JH, Kang HS, Kim JE, Chae JH, et al. Decreased level and defective function of circulating endothelial progenitor cells in children with moyamoya disease. J Neurosci Res. 2010; 88:510–518. PMID: 19774676.

32. Kim SK, Yoo JI, Cho BK, Hong SJ, Kim YK, Moon JA, et al. Elevation of CRABP-I in the cerebrospinal fluid of patients with Moyamoya disease. Stroke. 2003; 34:2835–2841. PMID: 14605320.

33. Kim YI, Phi JH, Paeng JC, Choi H, Kim SK, Lee YS, et al. In vivo evaluation of angiogenic activity and its correlation with efficacy of indirect revascularization surgery in pediatric moyamoya disease. J Nucl Med. 2014; 55:1467–1472. PMID: 25060195.

34. Kirkham FJ, DeBaun MR. Stroke in children with sickle cell disease. Curr Treat Options Neurol. 2004; 6:357. PMID: 15279758.

35. Koss M, Scott RM, Irons MB, Smith ER, Ullrich NJ. Moyamoya syndrome associated with neurofibromatosis Type 1 : perioperative and long-term outcome after surgical revascularization. J Neurosurg Pediatr. 2013; 11:417–425. PMID: 23373626.

36. Kuroda S, Hashimoto N, Yoshimoto T, Iwasaki Y. Research Committee on Moyamoya Disease in Japan. Radiological findings, clinical course, and outcome in asymptomatic moyamoya disease : results of multicenter survey in Japan. Stroke. 2007; 38:1430–1435. PMID: 17395863.

37. Kuroda S, Ishikawa T, Houkin K, Nanba R, Hokari M, Iwasaki Y. Incidence and clinical features of disease progression in adult moyamoya disease. Stroke. 2005; 36:2148–2453. PMID: 16179571.

38. Kusaka N, Sugiu K, Tokunaga K, Katsumata A, Nishida A, Namba K, et al. Enhanced brain angiogenesis in chronic cerebral hypoperfusion after administration of plasmid human vascular endothelial growth factor in combination with indirect vasoreconstructive surgery. J Neurosurg. 2005; 103:882–890. PMID: 16304993.

39. Lee JY, Kim SK, Cheon JE, Choi JW, Phi JH, Kim IO, et al. Posterior cerebral artery involvement in moyamoya disease : initial infarction and angle between PCA and basilar artery. Childs Nerv Syst. 2013; 29:2263–2269. PMID: 23653141.

40. Lin N, Baird L, Koss M, Kopecky KE, Gone E, Ullrich NJ, et al. Discovery of asymptomatic moyamoya arteriopathy in pediatric syndromic populations : radiographic and clinical progression. Neurosurg Focus. 2011; 31:E6.
41. Liu W, Morito D, Takashima S, Mineharu Y, Kobayashi H, Hitomi T, et al. Identification of RNF213 as a susceptibility gene for moyamoya disease and its possible role in vascular development. PLoS One. 2011; 6:e22542. PMID: 21799892.

42. Malek AM, Connors S, Robertson RL, Folkman J, Scott RM. Elevation of cerebrospinal fluid levels of basic fibroblast growth factor in moyamoya and central nervous system disorders. Pediatr Neurosurg. 1997; 27:182–189. PMID: 9577971.

43. Maruwaka M, Yoshikawa K, Okamoto S, Araki Y, Sumitomo M, Kawamura A, et al. Biomarker research for moyamoya disease in cerebrospinal fluid using surface-enhanced laser desorption/ionization time-of-flight mass spectrometry. J Stroke Cerebrovasc Dis. 2015; 24:104–111. PMID: 25440344.

44. Masuda J, Ogata J, Yutani C. Smooth muscle cell proliferation and localization of macrophages and T cells in the occlusive intracranial major arteries in moyamoya disease. Stroke. 1993; 24:1960–1967. PMID: 7902623.

45. Mineharu Y, Liu W, Inoue K, Matsuura N, Inoue S, Takenaka K, et al. Autosomal dominant moyamoya disease maps to chromosome 17q25.3. Neurology. 2008; 70(24 Pt 2):2357–2363. PMID: 18463369.

46. Miyatake S, Miyake N, Touho H, Nishimura-Tadaki A, Kondo Y, Okada I, et al. Homozygous c.14576G>A variant of RNF213 predicts early-onset and severe form of moyamoya disease. Neurology. 2012; 78:803–810. PMID: 22377813.

47. Moftakhar P, Smith ER, Choulakian A, Scott RM, Danielpour M. Moyamoya disease in children with congenital dwarfing conditions. Pediatr Neurosurg. 2010; 46:373–380. PMID: 21389750.

48. Munot P, Saunders DE, Milewicz DM, Regalado ES, Ostergaard JR, Braun KP, et al. A novel distinctive cerebrovascular phenotype is associated with heterozygous Arg179 ACTA2 mutations. Brain. 2012; 135(Pt 8):2506–2514. PMID: 22831780.

49. Nanba R, Kuroda S, Ishikawa T, Houkin K, Iwasaki Y. Increased expression of hepatocyte growth factor in cerebrospinal fluid and intracranial artery in moyamoya disease. Stroke. 2004; 35:2837–2842. PMID: 15528455.

50. Ni G, Liu W, Huang X, Zhu S, Yue X, Chen Z, et al. Increased levels of circulating SDF-1α and CD34+ CXCR4+ cells in patients with moyamoya disease. Eur J Neurol. 2011; 18:1304–1309. PMID: 21435112.

51. Qaiser R, Scott RM, Smith ER. Identification of an association between Robinow syndrome and moyamoya. Pediatr Neurosurg. 2009; 45:69–72. PMID: 19258733.

52. Qureshi IA, Mehler MF. Emerging role of epigenetics in stroke : part 1 : DNA methylation and chromatin modifications. Arch Neurol. 2010; 67:1316–1322. PMID: 21060009.
53. Qureshi IA, Mehler MF. The emerging role of epigenetics in stroke : II. RNA regulatory circuitry. Arch Neurol. 2010; 67:1435–1441. PMID: 21149808.
54. Roach ES, Golomb MR, Adams R, Biller J, Daniels S, Deveber G, et al. Management of stroke in infants and children : a scientific statement from a Special Writing Group of the American Heart Association Stroke Council and the Council on Cardiovascular Disease in the Young. Stroke. 2008; 39:2644–2691. PMID: 18635845.
55. Roder C, Peters V, Kasuya H, Nishizawa T, Wakita S, Berg D, et al. Analysis of ACTA2 in European Moyamoya disease patients. Eur J Paediatr Neurol. 2011; 15:117–122. PMID: 20970362.

56. Romeo MJ, Espina V, Lowenthal M, Espina BH, Petricoin EF 3rd, Liotta LA. CSF proteome : a protein repository for potential biomarker identification. Expert Rev Proteomics. 2005; 2:57–70. PMID: 15966853.

57. Sakurai K, Horiuchi Y, Ikeda H, Ikezaki K, Yoshimoto T, Fukui M, et al. A novel susceptibility locus for moyamoya disease on chromosome 8q23. J Hum Genet. 2004; 49:278–281. PMID: 15362573.

58. Scott RM, Smith ER. Moyamoya disease and moyamoya syndrome. N Engl J Med. 2009; 360:1226–1237. PMID: 19297575.

59. Scott RM, Smith JL, Robertson RL, Madsen JR, Soriano SG, Rockoff MA. Long-term outcome in children with moyamoya syndrome after cranial revascularization by pial synangiosis. J Neurosurg. 2004; 100(2 Suppl Pediatrics):142–149. PMID: 14758941.

60. Shimojima K, Yamamoto T. ACTA2 is not a major disease-causing gene for moyamoya disease. J Hum Genet. 2009; 54:687–688. PMID: 19745835.

61. Smith ER, McClain CD, Heeney M, Scott RM. Pial synangiosis in patients with moyamoya syndrome and sickle cell anemia : perioperative management and surgical outcome. Neurosurg Focus. 2009; 26:E10. PMID: 19335126.
62. Smith ER, Scott RM. Progression of disease in unilateral moyamoya syndrome. Neurosurg Focus. 2008; 24:E17. PMID: 18275294.

63. Smith ER, Scott RM. Spontaneous occlusion of the circle of Willis in children : pediatric moyamoya summary with proposed evidence-based practice guidelines. A review. J Neurosurg Pediatr. 2012; 9:353–360. PMID: 22462697.

64. Smith JJ, Sorensen AG, Thrall JH. Biomarkers in imaging : realizing radiology's future. Radiology. 2003; 227:633–638. PMID: 12663828.
65. Soriano SG, Cowan DB, Proctor MR, Scott RM. Levels of soluble adhesion molecules are elevated in the cerebrospinal fluid of children with moyamoya syndrome. Neurosurgery. 2002; 50:544–549. PMID: 11841722.

67. Sugiyama T, Kuroda S, Nakayama N, Tanaka S, Houkin K. Bone marrow-derived endothelial progenitor cells participate in the initiation of moyamoya disease. Neurol Med Chir (Tokyo). 2011; 51:767–773. PMID: 22123479.

68. Suzuki J, Takaku A. Cerebrovascular "moyamoya" disease. Disease showing
abnormal net-like vessels in base of brain. Arch Neurol. 1969; 20:288–299. PMID: 5775283.
69. Takahashi A, Sawamura Y, Houkin K, Kamiyama H, Abe H. The cerebrospinal fluid in patients with moyamoya disease (spontaneous occlusion of the circle of Willis) contains high level of basic fibroblast growth factor. Neurosci Lett. 1993; 160:214–216. PMID: 8247356.

70. Takeuchi K, Shimizu K. Hypoplasia of the bilateral internal carotid arteries. No To Shinkei. 1957; 9:37–43.
71. Ullrich NJ, Robertson R, Kinnamon DD, Scott RM, Kieran MW, Turner CD, et al. Moyamoya following cranial irradiation for primary brain tumors in children. Neurology. 2007; 68:932–938. PMID: 17372129.

72. Yamada M, Fujii K, Fukui M. [Clinical features and outcomes in patients with asymptomatic moyamoya disease--from the results of nation-wide questionnaire survey]. No Shinkei Geka. 2005; 33:337–342. PMID: 15830539.
73. Yanagawa Y, Sugiura T, Suzuki K, Okada Y. Moyamoya disease associated with positive findings for rheumatoid factor and myeloperoxidase-anti-neutrophil cytoplasmic antibody. West Indian Med J. 2007; 56:282–284. PMID: 18072414.

74. Yoshihara T, Taguchi A, Matsuyama T, Shimizu Y, Kikuchi-Taura A, Soma T, et al. Increase in circulating CD34-positive cells in patients with angiographic evidence of moyamoya-like vessels. J Cereb Blood Flow Metab. 2008; 28:1086–1089. PMID: 18231114.

75. Yoshimoto T, Houkin K, Takahashi A, Abe H. Angiogenic factors in moyamoya disease. Stroke. 1996; 27:2160–2165. PMID: 8969773.

Table 4
Molecular biomarkers of moyamoya
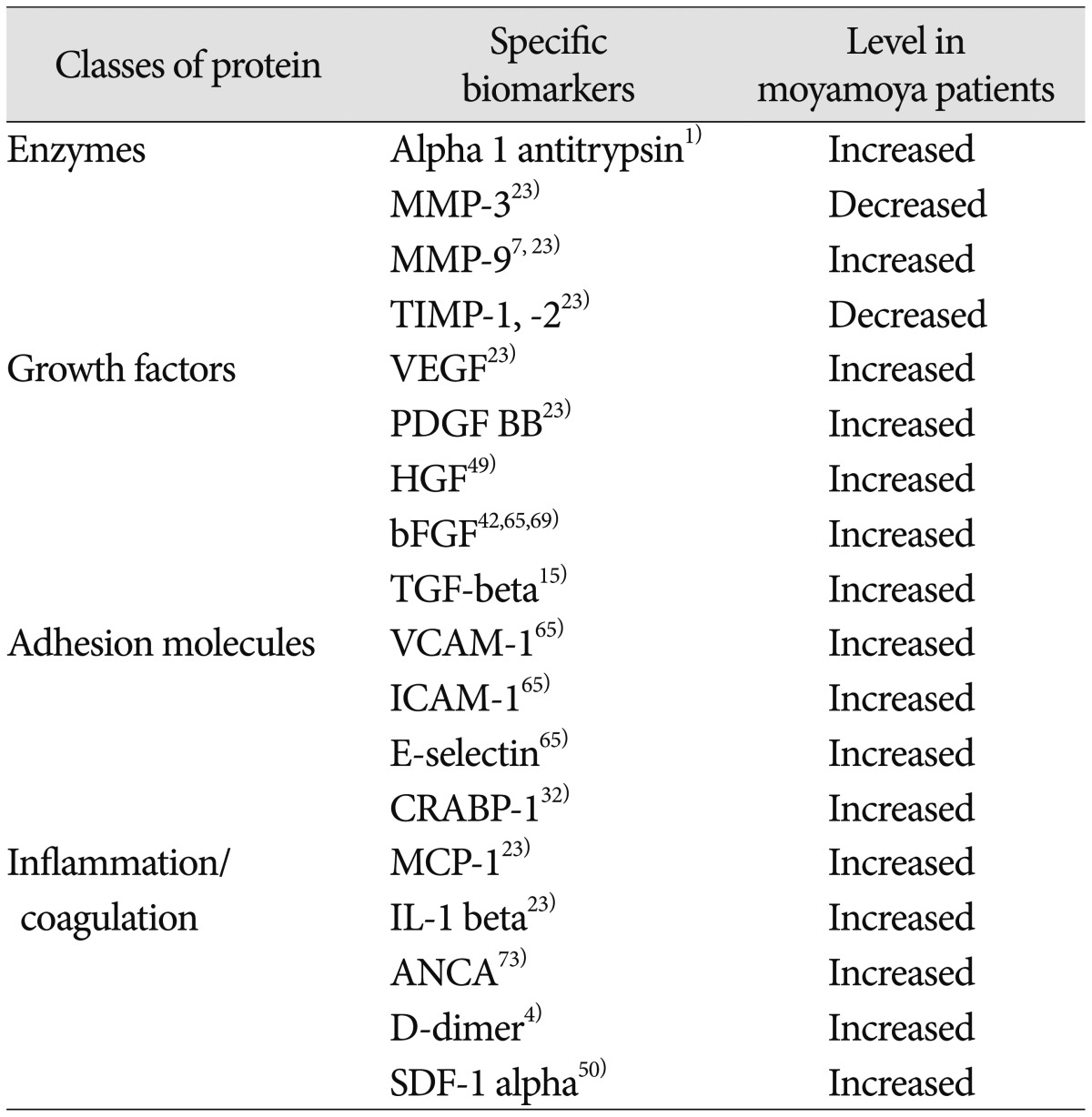
MMP : matrix metalloproteinase, TIMP : tissue inhibitor of metalloproteinase, VEGF : vascular endothelial growth factor, PDGF : platelet-derived growth factor, HGF : hepatocyte growth factor, bFGF : basic fibroblast growth factor, TGF-beta : transforming growth factor beta, VCAM-1 : vascular cell adhesion molecule 1, ICAM-1 : intercellular adhesion molecule 1, CRABP-1 : cellular retinoic acid binding protein 1, MCP-1 : monocyte chemotactic protein-1, IL-1 beta : interleukin 1 beta, ANCA : anti-neutrophil cytoplasmic antibodies, SDF-1 alpha : stromal cell-derived factor 1




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download