Abstract
Objective
The function of B7H3, a member of the B7 family of proteins, in neuroblastoma (NB) remains poorly characterized. Here we examine the expression pattern of B7H3 in clinical NB specimens and characterize the phenotype of B7H3 knock-down in NB cell line.
Methods
Immunohistochemical (IHC) staining was carried out to assess the expression of B7H3 in clinical NB specimens. Survival association was analyzed using five Gene Expression Omnibus (GEO) datasets (GSE85047, GSE45480, GSE62564, GSE16476, GSE49710). Clonogenic survival and flow cytometry were performed after B7H3 knockdown to assess the cellular proliferation and cell survival in vitro. Impact of B7H3 silencing on NB growth was examined in vivo using the SH-SY5Y xenograft model.
Results
On IHC staining, B7H3 was widely expressed in clinical NB specimens. Analysis of the transcriptional profiles of five GEO datasets clinically annotated NB specimens revealed that decreased B7H3 expression was associated with improved overall survival. B7H3 knockdown suppressed the proliferation of the SH-SY5Y NB model in vitro and in vivo. Cell cycle analysis revealed that B7H3 silencing induced G1/S arrest. This arrest was associated with the suppression of E2F1 expression and induction of Rb expression.
Neuroblastoma (NB) is the most common extracranial solid tumor of childhood, with 25–50 cases per million of individuals. It is a neuro-endocrine tumor that arise from the developing neural crest elements, resulting in tumors in the sympathetic ganglia or adrenal glands [2,11] Several driver genetic alterations have been found in NB, including N-myc protooncogene protein amplification (20%), ALK receptor tyrosine kinase mutation (8–10%), and telomerase reverse transcriptase rearrangement (31%) [5,14]. Despite the critical importance of these onco-proteins, the published literature suggest a wider range of targets available for NB therapeutic development [4,16,20]. Here we examined B7H3 as such a target.
B7H3 is a member of the B7 superfamily, a group of proteins that modulate T-cell response through cell to cell interaction [2,6]. Most published studies have focused on the immune-modulatory function of B7H3. Recently, studies demonstrate that B7H3 expression could promote cell migration and invasion in various cancer, including melanoma [17], breast cancer [9], colorectal cancer [3], prostate cancer [19], and gastric cancer [3]. Our previous works reveal the underlying mechanism of B7H3 in regulating tumor differentiation of glioblastoma cells by modulating transforming growth factor β pathway [20]. Besides, it has been shown that B7H3 also reprogram metabolic progress [8,10]. Coexpression of IDH1 and B7H3 can predict poor survival in colorectal cancer patients [18]. In spite of the lack of receptor, B7H3 has been demonstrated as an effective therapeutic target against tumor vascular cells angiogenesis [15]. However, there has been little focus on the influence of B7H3 on cell intrinsic processes in NB, such as cell cycle progression. And the precise molecular mechanisms of B7H3 in mediating NB tumor has never delineated before.
In this study, to figure out the specific function of B7H3 in NB tumor, we explored the expression pattern and prognosis value of B7H3 in clinical NB specimens, and examine the phenotype of B7H3 knock-down in NB cell line and xenograft model.
Human NB tissue samples (obtained from Children’s Hospital of Fudan University with full consent after approval from Local Ethics Committee, No. 2017-66) were fixed with 4% formaldehyde. Paraffin-embedded tumor tissues were sectioned to 5 µm thickness and mounted on positively charged microscope slides. And 1 mM ethylene diamine tetraacetic acid (pH 8.0) for B7H3 was used for antigen retrieval. Endogenous peroxidase activity was quenched by incubating the slides in methanol containing 3% hydrogen peroxide, followed by washing in phosphate buffered saline (PBS) for 15 minutes. The sections were incubated for 30 minutes at 37℃ with normal goat serum and subsequently incubated at 4℃ overnight with primary B7H3 antibodies (1 : 200, ab134161; Abcam, Cambridge, MA, USA). The sections were then rinsed with PBS and incubated with horseradish peroxidase-conjugated goat anti-mouse antibodies, followed by reaction with diaminobenzidine and counterstaining with Mayer's hematoxylin. The Immunoreactive Score (IRS) was used for B7H3 IHC evaluation and score from 0 to 12 points was obtained by two separate pathologists. Samples with IRS values between 0 to 2 were considered negative, 3 to 5 were positive (+), 6 to 8 were moderate positive (++), 9 to 12 were strong positive (+++).
The five microarray expression profiles (GSE85047, GSE45480, GSE62564, GSE16476, GSE49710) was extracted from the Gene Expression Omnibus (GEO) database from R2 : Genomics Analysis and Visualization Platform (http://r2.amc.nl). A total of 1843 NB samples in five GEO series were available for this study. After obtaining the raw data, the robust multiarray average method of the linear models for microarray data package in bioconductor was used to perform background correction, quartile normalization and make summarization. The optimal cutoff point with maximal sensitivity and specificity was calculated by ‘SurvivalROC’ package. Based on the optimal cutoff point, the patients in the datasets were divided into a low-expression and a high-expression group. And overall survival (OS) curves were plotted and used to analyze the prognostic impact based on above classifier.
Human NB cell line (SH-SY5Y), which was a gift from Professor Liuguan Bian in Ruijin Hospital, Shanghai Jiao Tong University, was grown in DMEM medium (ThermoFisher, Waltham, MA, USA) supplemented with 10% fetal bovine serum (Gibco, Waltham, MA, USA), penicillin (100 U/mL), and streptomycin (100 µg/mL) at 37℃ containing 95% air and 5% CO2. The usage of gifted cell line was approved by the donor and the Institutional Ethics Committee of Children’s Hospital, Fudan University.
The gene expression level of B7H3 in NB tumor cell line sets were accessed at the cBioPortal for Cancer Genomics (www.cbioportal.org/). Scrambled shRNA (5'TTCTCCGAACGTGTCACGT-3') that does not target any genes was used as the negative control, and the sh1-B7H3 (5'-CAAAG AAG ATGATGGACAA GACTCGAGTCTTGTCCATCATCTTCTTTGTTTTTTG-3') and sh2-B7H3 (5'- CC GGCTCTGAAACACTCTGACAGCACTCGAGTGCTGTCAGAGTGTTTCAGAGTTTTTTG-3') were selected for shRNA to eliminate cross-silence phenomenon with non-target genes. shRNA was constructed by annealing the synthetic DNA oligonucleotide primers, then naturally cooled to room temperature, then the oligonucleotides encoding shRNA specific for B7H3 and scramble sequence were subcloned into AgeI and EcoRI restrictive site of PLKO.1 vector. The plasmids sh-B7H3 and sh-Scr were verified by DNA sequencing and transfected into SH-SY5Y cell line, and the stable tumor cells were screened by administration of puromycin (1 : 1000) for 1 week.
For colony formation analysis, the lentivirus-transduced cells were digested with trypsin, counted by Countstar Automated Cell Counter (Shanghai Rui Yu Biotechnology Company, Shanghai, China), and seeded in six-well plates at a density of 500 cells per well. The medium was changed every 7 days for 14 days until visible colonies formed. Colonies were fixed by 4% polyformaldehyde for 30 minutes and stained with crystal violet for 15 minutes, and colonies with more than 20 cells was counted.
For cell proliferation analysis, the lentivirus-transduced cells were digested with trypsin, counted by Countstar Automated Cell Counter (Shanghai RuiYu Biotechnology Company), and seeded in 6-well plates at a density of 105 cells per well.
The effect of B7H3 on cell cycle distribution was determined by flow cytometry. Briefly, the lentivirus-transduced cells (1×105 cells/dish) were seeded at 6-cm dishes with three replications. Cells were digested with trypsin when they reached 80% confluence, and fixed for at least 24 hours with 75% ice-cold ethanol at 4℃. Cells were washed with ice-cold PBS and resuspended in 500 mL mixed liquids containing 50 µL/mL propidium iodide and 100 µL/mL RNaseA (Cell Cycle and Apoptosis Analysis Kit, C1052; Beyotime Biotechnology, Shanghai, China). After following incubation for 30 minutes in the dark at 37℃, cells were analyzed by flow cytometry (Becton-Dickinson, San Jose, CA, USA). The fractions of cells in G0/G1, S, and G2/M phases were analyzed using ModFit LT software (Verity Software House, Topsham, ME, USA).
Stable cells were washed twice with PBS and suspended in a lysis buffer (2% Mercaptoethanol, 20% Glycerol, 4% SDS in 100 mM Tris-HCl buffer, pH 6.8). Equal amount of proteins were loaded and separated by SDS-PAGE, and then transferred onto PVDF membrane (Schleicher&Schuell Co, Keene, NH, USA) using an electro- blotting apparatus (Bio-RAD, Hercules, CA, USA). The membrane was blocked with 5% nonfat milk (R&D, Minneapolis, MN, USA) in TBST solution for 1 hour at room temperature, and incubated overnight at 4℃ with specific antibody to B7H3 (1 : 1000, ab134161; Abcam), cyclinD1 (1 : 1000, ab134175; Abcam), Rb (1 : 1000, ab181616; Abcam), E2F1 (1 : 1000, ab179445; Abcam). After three washes in TBST solution, the membrane was incubated with secondary antibody diluted with TBST solution at room temperature for 2 hours. The signals of detected proteins were visualized on Image Quant LAS 4000 mini detection system (GE, Boston, MA, USA). β-actin were used as a loading control.
Total RNA was isolated using Trizol (ThermoFisher, Waltham, MA, USA) according to the manufacturer’s instructions and 1ug of total RNA from each cell line was then transcribed into cDNA. Reverse transcription and PCR were performed using a Takara RNA PCR (AMV) (Takara, Kusatsu, Japan) kit with primers designed for murine genes. The primers were synthetized by Invitrogen. The results were normalized against the level of glyceraldehyde 3-phosphate dehydrogenase as the internal control. RT-PCR was performed in triplicate for each experiment, including for the non-template controls.
The anti-proliferation effect of B7H3 suppression was evaluated in vivo using the SH-SY5Y-bearing subcutaneous xenograft model in 3-week aged BALB/c nude mice (n=5 per group). The mice were obtained from Slac Laboratory Animal Company (Shanghai, China) and maintained under specific pathogen free condition. The experiments conformed to the Animal Management Rule of the Chinese Ministry of Health, and the experimental protocol was approved by the Animal Care and Use Committee of Fudan University. When cells reached the confluence of 80–90%, they were digested and suspended at a density of 1×108/mL PBS, and 0.1 mL were subcutaneously injected into the right lower abdomen location without anesthesia. The mice were separately housed in a temperature-controlled environment with a 12 hours light and 12 hours dark cycle (7 AM to 7 PM). Tumor size was measured every 7 days using microcaliper. The animals were sacrificed using cervical dislocation method, and tumors were excised after 21 days after injection. Tumor volume and weight were measured. Tumor volume was calculated according to the formula V = 1/2 ab2, with “a” representing the longest diameter and “b” representing the shortest diameter. Sections were cut from paraffin-embedded tissue, then antigen retrieval were performed by heating samples for 20 minutes at 95℃ in citrate buffer (pH 6.0). Sample were cooled to room temperature and incubated in 3% hydrogen peroxide to quench peroxidase activity. After being incubated at 4℃ overnight, the first batch of antibodies were washed with PBS, and biotin-labeled second antibody was added for 20 minutes at 37℃. After washing with PBS, diaminobenzidine and hematoxylin counterstaining was performed.
All data were presented as the mean±standard deviation. Data analysis was performed by one-way analysis of variance. For comparison of two groups, a student’s t-test was used. Differences with p<0.05 were considered to be statistically significant.
Animal studies was approved by the Animal Care and Use Committee of Fudan University. And the usage of NB tissue samples was approve by ethics committee of Children’s Hospital, Fudan University (reference number : 2017-66). The written informed consents were obtained from all participants or the legal guardian if the patient was under the age of 18 years. All procedures were performed in accordance with the guidelines and regulations as approved by Committee of Fudan University, which conformed to the principles of animal protection, and human ethics.
To examine B7H3 expression in clinical NB specimens, IHC staining was performed of NB. In all 18 specimens examined, positive expression of B7H3 was observed in 17 cases of NB specimens, including three cases in positive expression (+), nine cases in moderate positive expression (++), five cases in strong positive expression (+++) (Fig. 1A). Having confirmed that B7H3 is widely overexpressed in NB cells, we next assessed whether variation in B7H3 expression harbor prognostic implication in NB patients. To this end, we analyzed the RNA expression profiles of five GEO datasets including 1864 cases of NB specimen with clinical information. Kaplan-Meier analysis revealed that high B7H3 expression was associated with poor OS in all five datasets (p<0.05, Fig. 1B-F).
In this study, the NB cell line SH-SY5Y was chosen for further experiments. As a first step characterize the physiologic function of B7H3, we established stable knock down of B7H3 in SH-SY5Y. In the stable knock down line, the B7H3 mRNA and protein expression were decreased by 76±8.7% and 82±7.4%, respectively, relative to Sh-scrambled control (Fig. 2A).
We observed that the B7H3 knock down SH-SY5Y cell line exhibited a slower growth kinetic relative to the Shscrambled control. Three days after plating, the number of cells produced by the B7H3 silenced lines was reduced by 62.5±6.8% (p<0.01) relative to the Sh-scrambled control in cell proliferation assay (Fig. 2B). In addition, the colony number of the B7H3 silenced lines was reduced by 72.3±8.5% (p<0.01) relative to the Sh-scrambled control at 14 days after plating (Fig. 2C). Obviously, B7H3 knockdown could inhibit proliferation of NB cells in vitro.
We next determined whether B7H3 silencing affected cell cycle progression. When compared with the Sh-scrambled controls, the B7H3 knock down lines showed increased accumulation of cells at the G0/G1 phase relative to the Sh-scrambled control (77.20±0.47% vs. 55.66±0.73%, p<0.001) (Fig. 3A).
Rb, E2F1, and cyclinD1 play key roles in the regulation of the G1 transition [7,12]. We, therefore, examined whether B7H3 silencing affected the expression of these proteins. Notably, we observed that Sh-B7H3 suppress the expression of Cyclin D1 and E2F1 while inducing the expression of Rb compared to Sh-scrambled control (Fig. 3B). In the GSEA45480 dataset, the Pearson correlation coefficient between B7H3 and Rb was -0.430 (p<0.0001), B7H3 and cyclinD1 was 0.378 (p<0.0001), B7H3 and E2F1 was 0.170 (p<0.0001) (Fig. 3C). These results suggested that B7H3 expression regulate the G1-S transition through simultaneous modulation of Rb, E2F1, and CyclinD1.
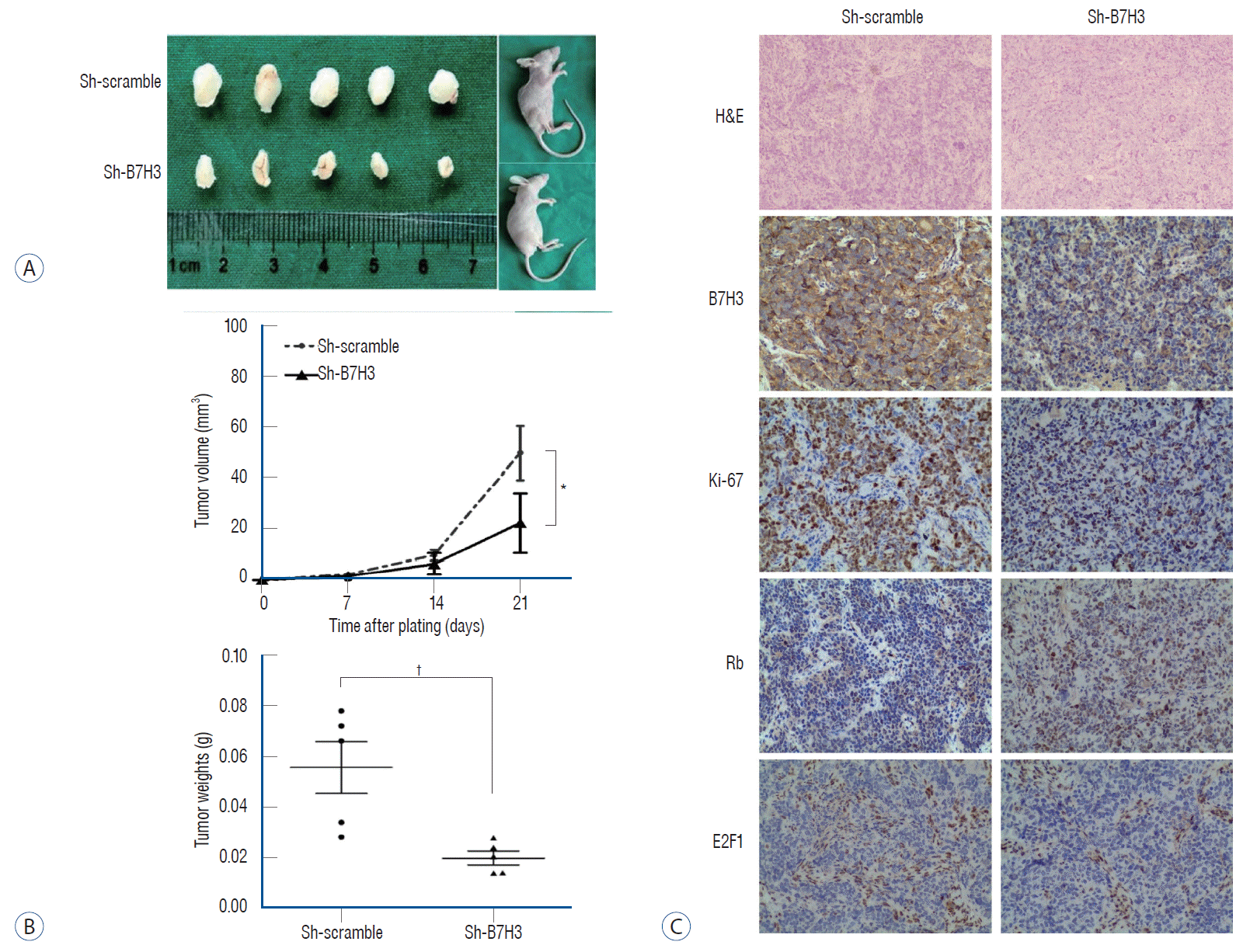
We wished to validate our in vitro and clinical observations using an in vivo xenograft model. Since the SH-SY5Y cell line for subcutaneous xenografts, we compared the growth kinetics of the Sh-B7H3 SH-SY5Y line relative to the Sh-scrambled SH-SY5Y line. The growth of the B7H3 knockdown line was significantly slowed relative to the Sh-scrambled line (Fig. 4A). The tumor volumes and weights in the Sh-B7H3 group were significantly smaller than that in the control group on day 21 after transplantation (tumor volume : 22.23±11.31 vs. 50.42±10.72 mm3, p<0.05; tumor weight : 0.02±0.00 vs. 0.06±0.01 g, p<0.01) (Fig. 4B).
We next determined whether this slowed growth in vivo was associated with up-regulation of Rb as well as the suppression of E2F1. IHC staining was performed using harvested xenograft tumor. These analyses confirmed our in vitro observation. In the xenografts, B7H3 knockdown was associated with lower Ki-67 positive staining ratio (42.67±6.43% vs. 69.00±14.93%, p<0.05), lower E2F1 expression (IRS score : 3.33±0.58 vs. 5.33±1.15, p<0.05) and higher Rb expression (IRS score : 6.33±2.52 vs. 2.67±1.15, p<0.05) (Fig. 4C), which is consistent with data in vitro. These results provide support of a model that B7H3 was associated with up-regulation of Rb as well as the suppression of E2F1.
The role of B7H3 in NB remains poorly defined. Here, we demonstrate that B7H3 is highly expressed in clinical NB specimens. Moreover, expression analysis revealed significant association between B7H3 expression and OS in five. These results suggest that B7H3 play an important role in NB biology. Silencing of B7H3 suppressed NB growth. This growth arrest is associated with compromised G1/S transition, suppression of E2F1 and induction of Rb. These results suggest that B7H3 mediate tumor cell-intrinsic physiology (e.g., cell cycle, progression) in addition to cell-extrinsic processes (e.g., immune regulation).
The mechanism through which B7H3 mediate Rb and E2F expression will require further molecular dissection. Rb and E2F are important for cell cycle [1,13], which partially explained the effect of B7H3 on cell proliferation. However, only co-expression of B7H3 and Rb/E2F could be observed according to our data now. Challenges in addressing the mechanistic link between B7H3 and Rb/E2F is compounded by the poorly understood cell-intrinsic properties of B7H3 and the undefined cognate ligand.
The association of B7H3 and cell apoptosis has been reported in other cancers. For example, B7H3 could inhibit apoptosis through Jak2-STAT3 pathway in colorectal cancer [21]. However, in our study, whether the proliferation arrest by silencing of B7H3 is caused by the pro-apoptosis function, further studies need to be performed in future.
In conclusion, our results indicated that B7H3 is highly expressed in human NB. Moreover, B7H3 expression is tightly associated with patients OS rate. Our result indicate that B7H3 is an essential regulator of NB cell cycle progression and represent a potential target for the development of NB therapy.
Notes
ACKNOWLEDGMENTS
This study was supported by National Natural Science Foundation of China (Grand No. 81572483 and 81611130223), Program of International Science & Technology Cooperation of China (Grand No. 2014DFA31470), Shanghai Committee of Science and Technology, China (No.15441904500) and Natural Science Foundation and Major Basic Research Program of Shanghai, China (No. 16JC1420100). CCC is supported by 1RO1NS097649-01, the Doris Duke Charitable Foundation Clinical Scientist Development Award, The Sontag Foundation Distinguished Scientist Award, the Kimmel Scholar Award, and BWF 1006774.01. These funders support authors in finance, not participating in the experimental or writing works.
References
1. Bertoli C, Skotheim JM, de Bruin RA. Control of cell cycle transcription during G1 and S phases. Nat Rev Mol Cell Biol. 14:518–528. 2013.

2. Castriconi R, Dondero A, Augugliaro R, Cantoni C, Carnemolla B, Sementa AR, et al. Identification of 4Ig-B7-H3 as a neuroblastoma-associated molecule that exerts a protective role from an NK cell-mediated lysis. Proc Natl Acad Sci U S A. 101:12640–12645. 2004.

3. Dong P, Xiong Y, Yue J, Hanley SJB, Watari H. B7H3 as a promoter of metastasis and promising therapeutic target. Front Oncol. 8:264. 2018.

4. Du H, Hirabayashi K, Ahn S, Kren NP, Montgomery SA, Wang X, et al. Antitumor responses in the absence of toxicity in solid tumors by targeting B7-H3 via chimeric antigen receptor T cells. Cancer Cell. 35:221–237.e8. 2019.

5. Eleveld TF, Oldridge DA, Bernard V, Koster J, Colmet Daage L, Diskin SJ, et al. Relapsed neuroblastomas show frequent RAS-MAPK pathway mutations. Nat Genet. 47:864–871. 2015.

6. Jeon H, Vigdorovich V, Garrett-Thomson SC, Janakiram M, Ramagopal UA, Abadi YM, et al. Structure and cancer immunotherapy of the B7 family member B7x. Cell Rep. 9:1089–1098. 2014.

7. John RR, Malathi N, Ravindran C, Anandan S. Mini review: multifaceted role played by cyclin D1 in tumor behavior. Indian J Dent Res. 28:187–192. 2017.

8. Lim S, Liu H, Madeira da Silva L, Arora R, Liu Z, Phillips JB, et al. Immunoregulatory protein B7-H3 reprograms glucose metabolism in cancer cells by ROS-mediated stabilization of HIF1α. Cancer Res. 76:2231–2242. 2016.

9. Liu H, Tekle C, Chen YW, Kristian A, Zhao Y, Zhou M, et al. B7-H3 silencing increases paclitaxel sensitivity by abrogating Jak2/Stat3 phosphorylation. Mol Cancer Ther. 10:960–971. 2011.

10. Luo D, Xiao H, Dong J, Li Y, Feng G, Cui M, et al. B7-H3 regulates lipid metabolism of lung cancer through SREBP1-mediated expression of FASN. Biochem Biophys Res Commun. 482:1246–1251. 2017.

12. McNair C, Xu K, Mandigo AC, Benelli M, Leiby B, Rodrigues D, et al. Differential impact of RB status on E2F1 reprogramming in human cancer. J Clin Invest. 128:341–358. 2018.

13. Otto T, Sicinski P. Cell cycle proteins as promising targets in cancer therapy. Nat Rev Cancer. 17:93–115. 2017.

14. Peifer M, Hertwig F, Roels F, Dreidax D, Gartlgruber M, Menon R, et al. Telomerase activation by genomic rearrangements in high-risk neuroblastoma. Nature. 526:700–704. 2015.

15. Seaman S, Zhu Z, Saha S, Zhang XM, Yang MY, Hilton MB, et al. Eradication of tumors through simultaneous ablation of CD276/B7-H3-positive tumor cells and tumor vasculature. Cancer Cell. 31:501–515. e8. 2017.

16. Tan WQ, Chen G, Ye M, Jia B. Artemether regulates chemosensitivity to doxorubicin via regulation of B7-H3 in human neuroblastoma cells. Med Sci Monit. 23:4252–4259. 2017.

17. Tekle C, Nygren MK, Chen YW, Dybsjord I, Nesland JM, Maelandsmo GM, et al. B7-H3 contributes to the metastatic capacity of melanoma cells by modulation of known metastasis-associated genes. Int J Cancer. 130:2282–2290. 2012.

18. Wu J, Wang F, Liu X, Zhang T, Liu F, Ge X, et al. Correlation of IDH1 and B7H3 expression with prognosis of CRC patients. Eur J Surg Oncol. 44:1254–1260. 2018.
19. Yuan H, Wei X, Zhang G, Li C, Zhang X, Hou J. B7-H3 over expression in prostate cancer promotes tumor cell progression. J Urol. 186:1093–1099. 2011.

Fig. 1.
B7H3 was associated with neuroblastoma prognosis. A : IHC staining showed that B7H3 was widely expressed in human NBs. In 18 patients, 17 cases had positive B7H3 expression, including three cases in positive expression (+), nine cases in moderate positive expression (++), five cases in strong positive expression (+++). B : High expression of B7H3 had worse overall survival than those with low expression in GSE85047 (n=283, p=2.7×10-5). C : High expression of B7H3 had worse overall survival than those with low expression in GSE62564 (n=498, p=1.1×10-3). D : High expression of B7H3 had worse overall survival than those with low expression in GSE45480 (n=476, p=7.5×10-4). E : High expression of B7H3 had worse overall survival than those with low expression in GSE16476 (n=88, p=1.0×10-3). F : High expression of B7H3 had worse overall survival than those with low expression in GSE49710 (n=498, p=1.7×10-2). IHC : immumohistochemistry, NB : neuroblastoma.
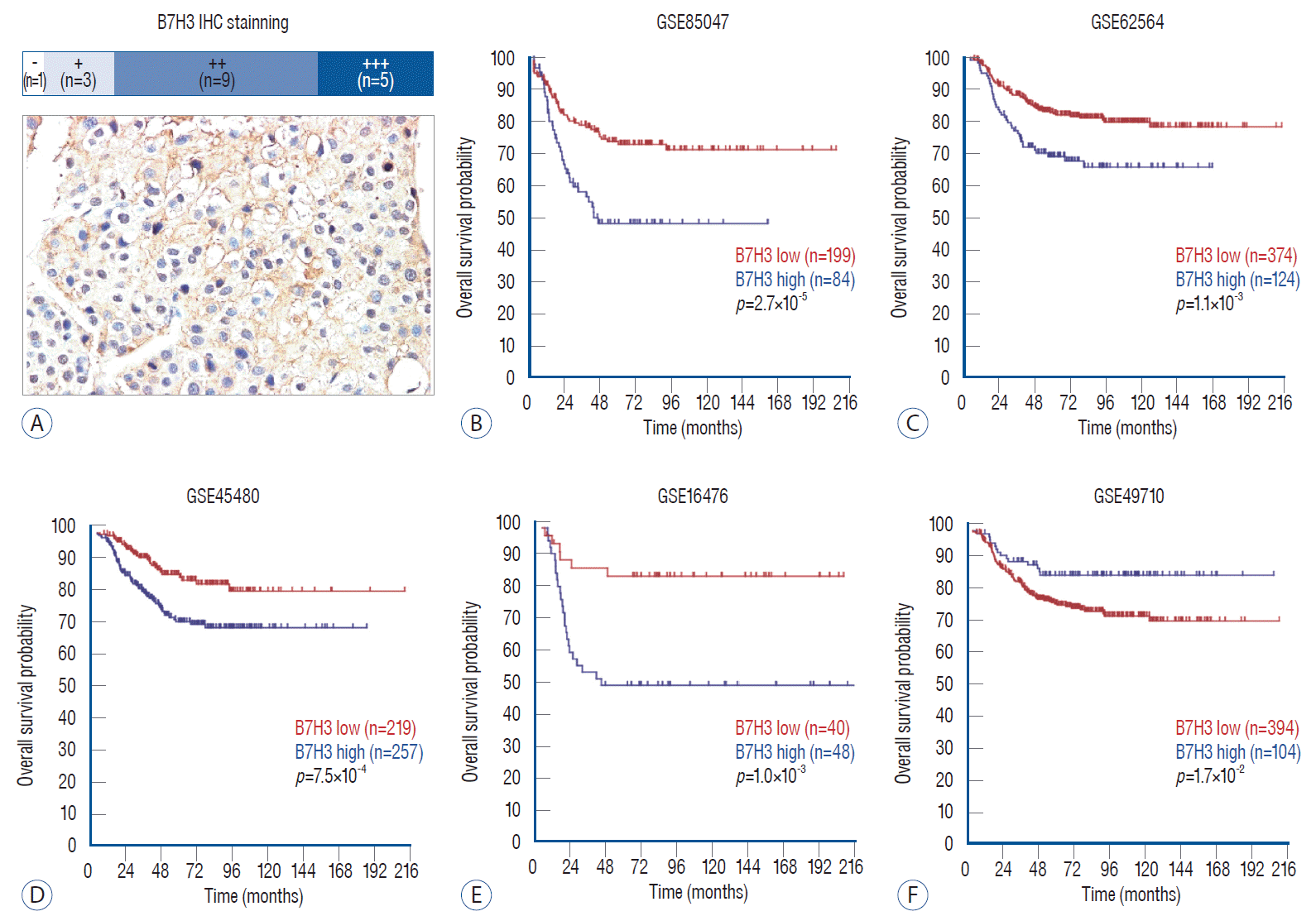
Fig. 2.
B7H3 knockdown could inhibit proliferation of neuroblastoma in vitro. A : B7H3 mRNA and protein level was effectively inhibited by shRNAs than control group in SH-SY5Y cell line. B : Cell proliferation was significantly inhibited in B7H3 knockdown group in SH-SY5Y cell line at the third day after plating (p<0.01). C : Colony numbers in B7H3 knockdown group in SH-SY5Y cell line was significantly reduced by 72.3±8.5% (p<0.001). *p<0.05. †p<0.01. GAPDH : glyceraldehyde 3-phosphate dehydrogenase, NS : not significant.
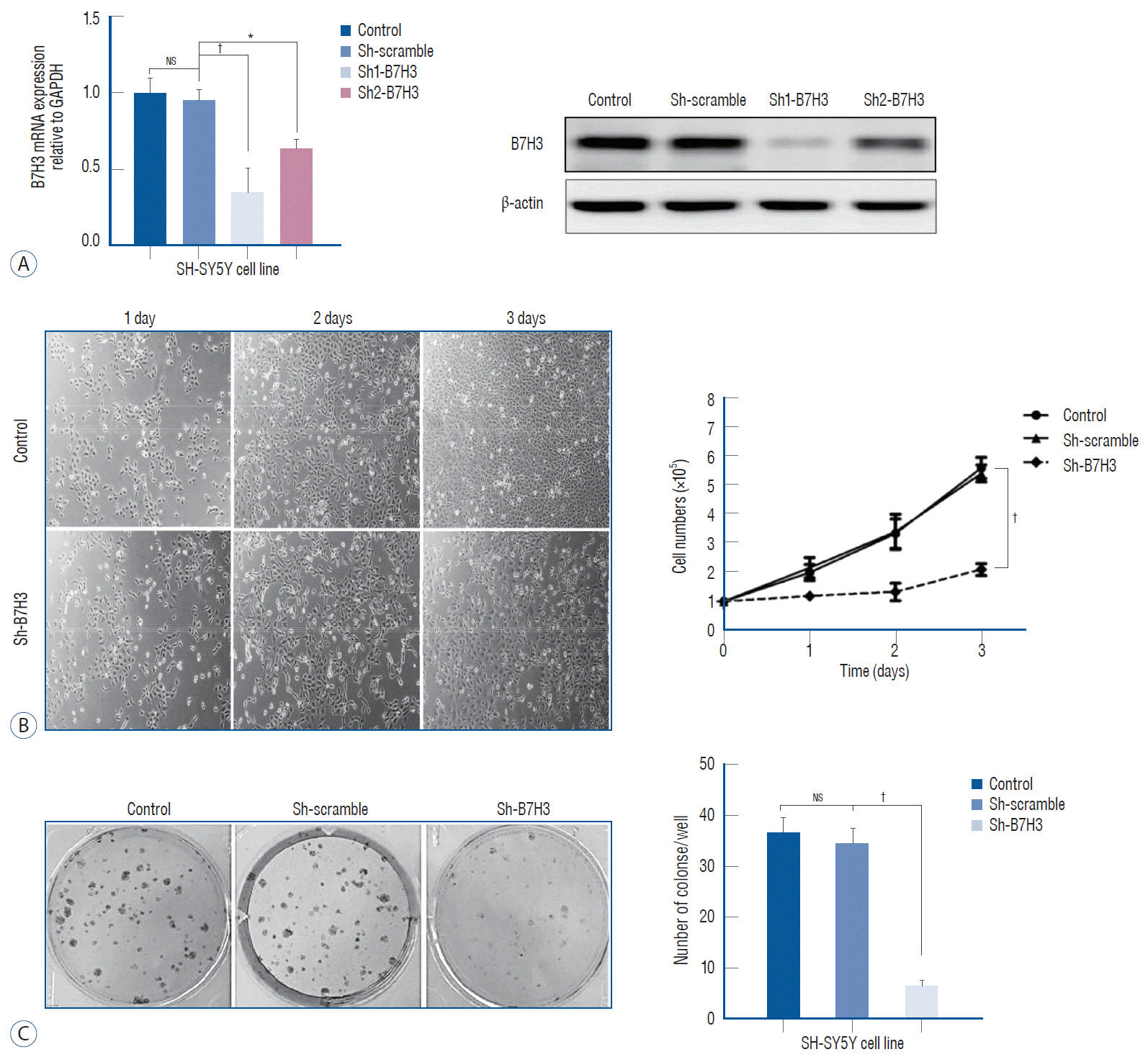
Fig. 3.
Cell cycle was suppressed in G1/S phase with checkpoint change after B7H3 knockdown. A : B7H3 knockdown inhibit G1/S phase transition, with G1/S : 56.22%/35.16% in control group and G1/S : 76.89%/19.73% in B7H3 knockdown group in SH-SY5Y cell line. B : RT-PCR (left) and western blot (middle and right) analysis of G1 phase checkpoint (cyclinD1, Rb, E2F1) in SH-SY5Y cell line. Relative protein expression values were normalized assigning the value of the cells in control groups to 1.0. C : Bioinformatic analysis of the correlation between B7H3 and proliferation-associated molecules. *p<0.01. †p<0.001. GAPDH : glyceraldehyde 3-phosphate dehydrogenase.
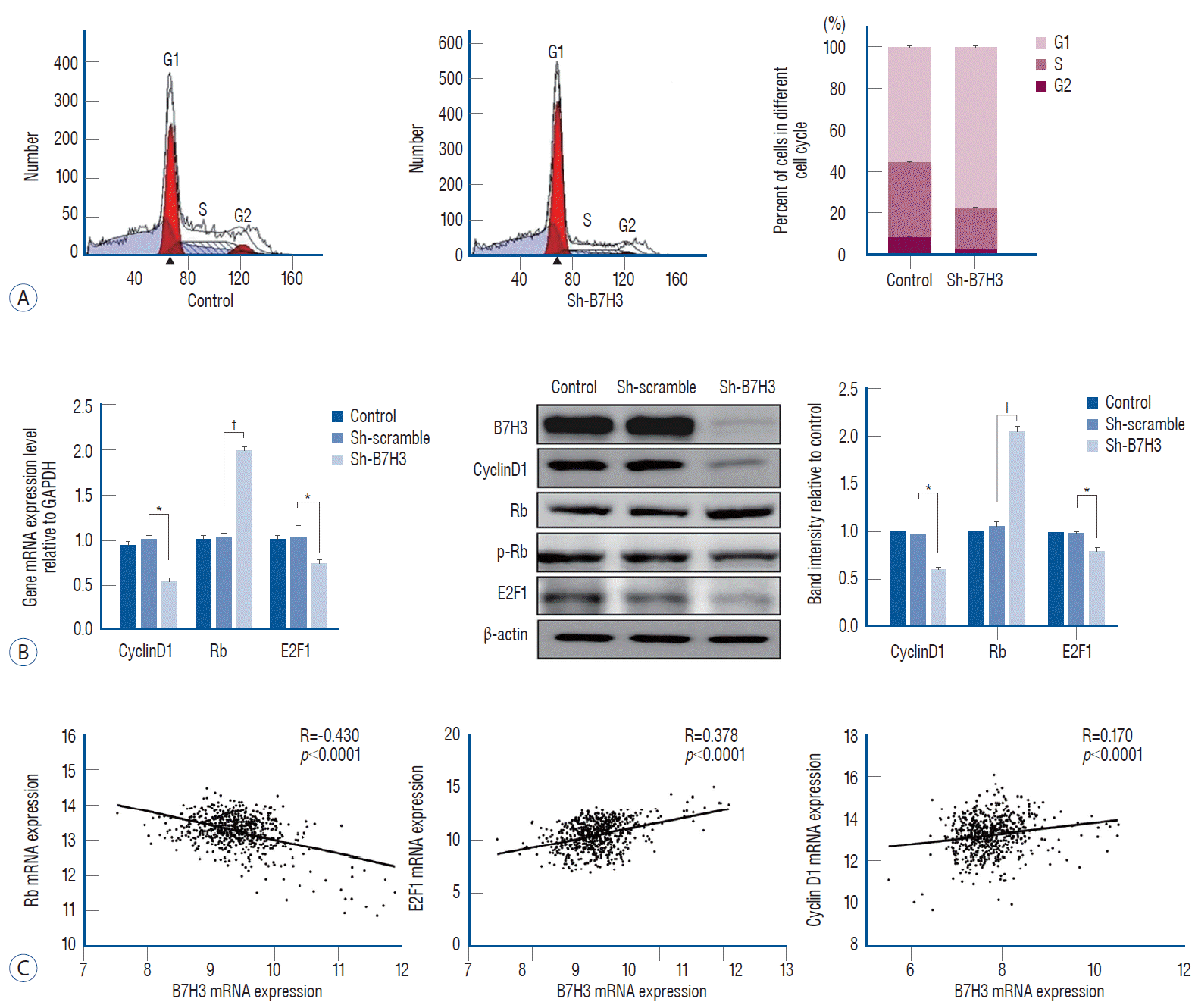
Fig. 4.
B7H3 knockdown could inhibit proliferation of neuroblastoma in vivo. A : Subcutaneous xenograft tumor model demonstrated that B7H3 knockdown could inhibit tumor growth. B : B7H3 knockdown inhibit tumor growth (22.23±11.31 vs. 50.42±10.72 mm3, p<0.05) and decrease tumor weight (0.02±0.00 vs. 0.06±0.01 g, p<0.01). C : Immunohistochemical staining showed that B7H3 knockdown group manifested lower Ki-67 index, E2F1 expression and higher Rb expression (×200). *p<0.05. †p<0.01. H&E : hematoxylin-eosin staining.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download