Abstract
Objective
To generate synthetic spine magnetic resonance (MR) images from spine computed tomography (CT) using generative adversarial networks (GANs), as well as to determine the similarities between synthesized and real MR images.
Methods
GANs were trained to transform spine CT image slices into spine magnetic resonance T2 weighted (MRT2) axial image slices by combining adversarial loss and voxel-wise loss. Experiments were performed using 280 pairs of lumbar spine CT scans and MRT2 images. The MRT2 images were then synthesized from 15 other spine CT scans. To evaluate whether the synthetic MR images were realistic, two radiologists, two spine surgeons, and two residents blindly classified the real and synthetic MRT2 images. Two experienced radiologists then evaluated the similarities between subdivisions of the real and synthetic MRT2 images. Quantitative analysis of the synthetic MRT2 images was performed using the mean absolute error (MAE) and peak signal-to-noise ratio (PSNR).
Results
The mean overall similarity of the synthetic MRT2 images evaluated by radiologists was 80.2%. In the blind classification of the real MRT2 images, the failure rate ranged from 0% to 40%. The MAE value of each image ranged from 13.75 to 34.24 pixels (mean, 21.19 pixels), and the PSNR of each image ranged from 61.96 to 68.16 dB (mean, 64.92 dB).
Conclusion
This was the first study to apply GANs to synthesize spine MR images from CT images. Despite the small dataset of 280 pairs, the synthetic MR images were relatively well implemented. Synthesis of medical images using GANs is a new paradigm of artificial intelligence application in medical imaging. We expect that synthesis of MR images from spine CT images using GANs will improve the diagnostic usefulness of CT. To better inform the clinical applications of this technique, further studies are needed involving a large dataset, a variety of pathologies, and other MR sequence of the lumbar spine.
Recently, remarkable advances in artificial intelligence (AI), especially deep learning, have been allowed the technology to be applied in medical image analysis. For example, convolution neural network (CNN), a class of deep learning algorithm, have shown remarkable performance in the classification of lesions on medical images [4,7,13]. Besides CNN, various other deep learning algorithms have been developed, and applied in the same context. Generative adversarial networks (GANs), which were introduced by Ian Goodfellow, have produced especially realistic images [5]. GANs have been used to synthesize positron emission tomography (PET) images from computed tomography (CT) images [2]. A study also has been reported to synthesize CT images from magnetic resonance (MR) images using GANs [12].
MR images and CT images are very important in the evaluation of lumbar spine diseases. In particular, CT scans are fast and suitable for bony structure analysis. However, they cannot distinguish soft tissues well. Conversely, MR scans are suitable for soft tissue evaluation, although they are sometimes contraindicated, such as in patients with claustrophobia or pacemakers. Moreover, MR scans are more expensive and require more time than CT scans.
Objective of the present study was to synthesize lumbar spine MR images from lumbar spine CT images using GANs. And, the similarities between synthesized and real MR images were quantitatively and qualitatively evaluated to confirm the feasibility of using this method in a clinical practice.
GANs can learn the way to synthesize MR images from CT images using mapping (G : ICT → IMR). The generator network, (G), is trained to generate realistic synthetic MR images that cannot be distinguished from “real” MR images by an adversarially trained discriminate network, D, which is trained to do as well as possible at detecting the generator’s “generated” (Fig. 1).
We applied adversarial losses to the generator network and its discriminator. The objective could be expressed as follows :
LGAN (G, D) = EICT, IMR - Pdata (ICT,IMR) [log D (ICT, IMR)] + EICT~Pdata (ICT) [log (1 - D (ICT, G(ICT)))]
whereby G tries to translate an ICT image to a G(ICT) image that looks similar to an image from the MR image domain. The discriminator D tries to discriminate between the real and synthesized pairs that provide ICT with synthesized MR image in the equation. The generator network G tries to minimize this objective against an adversarial D that tries to maximize it, i.e., G*= argmingmaxDLGAN(G, D).
Previous approaches have found it beneficial to combine the adversarial loss with a more traditional loss, such as L1 distance [14]. For the paired data (ICT, IMR), the generator network G is tasked to not only generate realistic MR images, but also to be near the reference IMR of the input ICT. The L1 loss term for the G was :
The overall objective was :
whereby λ control the relative importance of adversarial loss and voxel-wise loss.
After obtaining approval from Institutional Review Board of Pusan National University Hospital (1808-008-069), we collected CT and MR images from each patient who had undergone lumbar spine CT and MR scans within three days of each other. The CT scans were acquired on a 16-slice CT scanner (Revolution CT; GE Healthcare, Milwaukee, WI, USA). The MR images were acquired on a 1.5T MR scanner (Avanto; Siemens, Erlangen, Germany) and a 3T MR scanner (Skyra; Siemens). We then excluded CT and MR images of severe lumbar spine pathologies, such as tumor, infection or fracture, although we included images of degenerative disease. Among the MR images, magnetic resonance T2 weighted axial (MRT2) images were collected. Because this was a preliminary study to confirm the feasibility of GANs, only one type of MR sequence was selected. Among the CT and MRT2 images, we selected axial images that were parallel to the endplate of vertebral body and passed through the middle of the intervertebral disc. CT and MRT2 pairs that had different axes were excluded (Fig. 2). Two neurosurgeons selected CT and MRT2 images. To ensure efficient training, we augmented the training images. All images were adjusted in 256-grayscale. All real CT and MR images were cropped using following method. Horizontally, the image is cut parallel at the most ventral part of the lumbar vertebral body. It was then cut in the dorsal direction from the center of the thecal sac to the ventral end of the vertebral body. Finally, it was cut vertically by the same length at the center of the thecal sac. The range within which both lateral sides of the vertebral body can be seen was measured in all images (Fig. 3).
We obtained and reviewed lumbar spine CT and MR images performed at our hospital in 2017. Images conforming to the conditions mentioned above were confirmed in 129 patients (66 men, 63 women). The mean age of these patients was 61 years (range, 23–85). On average, 2.29 pairs of images (range, one to five pairs) were used per patient. A total 280 pairs of images were used as training data. Our algorithm then generated synthetic MRT2 images from 15 CT images other than the training images.
To create the generator network G, we used the architecture described by Johnson et al. [10], which is a 2D fully-convolutional network consisting of one convolutional layer followed by two strided convolutional layers, nine residual blocks, two fractionally-strided convolutional layers, and one last convolutional layer [6]. Instance normalization and ReLU followed all but the last convolution [15]. The synthesis network took a 256×256 input and generated an output image of the same size. For the discriminators D, we adapted PatchGANs, which classifies each N×N patch in an image as either real or fake [8]. In this way, the discriminators could focus better on high-frequency information in local image patches. Network D used two convolutions and five strided convolutions. Except for the first and last convolution, each convolutional layer was followed by instance normalization and leaky ReLu [15,17]. To optimize our networks, we used mini-batch stochastic gradient descent and applied the Adam optimizer with a batch size of 1 [11]. The learning rate started at 2e-4 for the first 1e5 iterations, and a decayed linearly to zero over the next 2e5 iterations. For all experiments, we set λ=10 empirically. At inference time, we ran the generator network G only to give a CT image.
The proposed approach training took about 20 hours for 2e5 iterations using a single GeForce GTX 1080Ti GPU. At inference time, the system required 35 ms to synthesize a single-slice CT image to MR image.
Real and synthesized MRT2 images were compared using the mean absolute error (MAE) :
where i is the index of the 2D axial image slice in aligned voxels, and N is the number of slices in the reference MRT2 images. The MAE measures the average distance between each pixel of the synthesized and the real MRT2 images. In addition, the synthesized MRT2 images were evaluated using the peak signal-to-noise ratio (PSNR) :
where MAX=255 [12]. PSNR measures the ratio between the maximum possible intensity value and the mean square error (MSE) of the synthesized and real MRT2 images. Smaller MSE values indicate more similarity between the two images. If there is no difference between two images, the MSE value is 0 and the PSNR value becomes infinite. In general, if the PSNR values >30 dB indicate that no differences can be distinguished by the human eye [18].
We made questionnaires showing synthetic and real MRT2 images corresponding to spine CT (Fig. 4). Six medical doctors who had never seen synthetic MRT2 images completed these questionnaires : two musculoskeletal radiologists, a senior spine surgeon, a junior spine surgeon, and two 4th-year neurosurgical residents. One of the radiologists and a senior spine surgeon had more than 15 years of experience. Other radiologists and a junior spine surgeon had about five years of experience. We then made another questionnaire to evaluate the similarity of each structure in the spine CT scans (Fig. 5). The following features were subdivided : disc signal, degree of disc protrusion, muscle, fat tissue, facet joint signal, degree of stenosis, thecal sac, bone, and overall similarity. The synthetic and real MRT2 images corresponding to the spine CT scan were shown simultaneously. Two radiologists then measured the similarity between the two MRT2 images as a percentage.
All CT images, the synthetic MRT2 images, and the real MRT2 images were shown in Fig. 6. The MAE values between the synthesized and real spine MRT2 images ranged from 13.74 to 34.24 pixels (Fig. 7). The PSNR value of all paired MR images were found to over 30 dB. Table 1 shows the MAE and PSNR values of each case.
In questionnaire to distinguish the real from the synthesized MRT2 image, the rates at which the synthetic image was chosen ranged from 0 % to 40 % (Table 2). The CT15 was the most frequently chosen case for the synthetic MRT2 image, even though the MAE of CT15 was 17.5 pixels, which was not the lowest.
The average overall similarity measured by the two radiologists was 80.2 % (Table 3). Image CT03 had the lowest overall similarity, while images CT01, CT02, CT12, and CT13 had highest overall similarity. Among all features, those with the highest similarity were muscle (87.5±6.3%) and fat tissue (86.3±12.5%), while disc signal (75.5±10%) and thecal sac (76.7±14.9%) had the lowest average similarity.
GANs are an emerging AI-based unsupervised learning technique that involves a pair of networks in competition with each other. Since their introduction in 2014, GANs have been applied in various areas, mainly image classification and regression of image, image synthesis, image-to-image translation, and super-resolution [3]. In the present study, we applied GANs to image synthesis and showed that synthetic systems can be trained, using paired data, to synthesize MRT2 images from CT scan. The approach utilized adversarial loss from a discriminator network, as well as voxel loss based on paired data, to synthesize realistic MR images. Quantitative evaluation showed that the synthesized MRT2 images were close approximations of the reference MRT2 image, achieving a PSNR>30 dB (Table 1).
In previous studies related to GANs, authors have used GANs to convert MR to CT images, or CT to PET images [2,12,16]. In particular, PET image synthesis can improve the accuracy of PET-based computer-aided diagnosis systems [2]. Studies converting MR to CT have reported that such techniques can prevent radiation exposure during CT scanning, as well as save the costs and time associated with additional imaging [12,16]. In the same studies, the synthetic images created using GANs were very similar to real images [2,9,12,16]. However, these results were obtained from quantitative analysis only; no qualitative analysis was carried out by clinicians or radiologist. In the present study, although the synthetic MRT2 images were quantitatively similar to the real MRT2 images, medical expert could not be deceived at all times. Although the MR images misrecognition rates of the neurosurgical resident and junior spine surgeon were relatively high, they did not exceed 50%.
The results of both quantitative and qualitative analysis differed between the synthetic MR and real MR images. A low MAE and high PSNR indicate quantitative similarity. Thus, cases CT04 and CT10 were the most similar. However, in the qualitative analysis, none of the six doctors misinterpreted these two cases as real MR images at all. The overall similarity was not rated as high by the radiologists either. In the qualitative analysis by physicians, CT15, CT12, and CT14 were misinterpreted as real MR images more frequently, while CT01, CT02, CT12, and CT13 had the highest similarity. Therefore, CT12 had the highest similarity in the qualitative analysis, but the second lowest in the quantitative analysis. As such, the qualitative and quantitative analyzes were discrepant. It follows that image conversion by AI should not be evaluated using quantitative methods only.
The structural similarity of each lumbar vertebrae between the CT and MR images, as measured by two radiologist, ranged from 40% to 100%. The features with the highest similarity were muscle and fat tissue, while the disc signal and thecal sac showed the least similarity between the synthesized and real images. These factors may have caused the difference between quantitative and qualitative similarity. In the MRT2 axial image, the area occupied by the paraspinal muscle was large, but that occupied by the thecal sac was small. In addition, the muscle has a simple structure, so the calculated quantitative similarity wound have been high. In contrast, neural structures and ligaments occupied a small area and thus wound not have contributed much to quantitative similarity. However, the radiologists focused more on these structures in the lumbar spine MRT2 images and may therefore have perceived a high qualitative difference, despite of the high quantitative similarity.
Because this was a preliminary study, there were some limitations. Firstly, no standard or criteria were used in the qualitative analysis. No previous research has evaluated similarity by comparing synthesized medical images to actual images. Thus, we devised these criteria ourselves; as such, they have not been verified. Future studies must establish criteria for similarity, and analysis by more radiologists is needed. Secondly, no severe pathologies were included in the images used for training or synthesis in the present study. Only relatively normal or simple degenerative lesions were included. Finally, only MRT2 axial images at disc level were synthesized because 1) in the case of degenerative diseases of lumbar spine, more information can be obtained from T2-weighted than T1-weighted images; 2) many degenerative diseases, such as spinal stenosis or disc protrusion, are visible at the disc level; and 3) among sagittal and coronal images it is difficult to find paired images because lordosis differs among individual shots. Thus, sagittal or coronal images must be reconstructed from synthetic axial MR images when required.
In the present study, the minimum overall average qualitative similarity measured by radiologists was 74.6%, while the maximum was 85.7%. These values are not satisfactory. Thus, synthetic MR images cannot completely replace actual MR images in the usual clinical practice. In particular, diseases with very low incidence, such as spinal tumors, may have a low similarity in synthetic MR images because of few numbers of training images [1]. Moreover, diagnosis or deciding treatment plan with synthetic images alone will lead to legal disputes. However, in special clinical conditions that CT scan is possible but MR scan is not possible, these synthetic MR images may increase the diagnostic usefulness of CT images. Since these were based on a relatively small dataset and had high quantitative similarity, they warrant further study including a large dataset, various pathologies, and other MR sequences of the lumbar spine.
This was the first study to apply GANs to synthesize spine MR images from CT images. Despite a small dataset of 280, the synthetic MR images were relatively well implemented. Synthetic medical imaging using GANs is a new AI paradigm in medical imaging. MR image synthesis using this method may improve the diagnostic usefulness of CT. To inform clinical applications, further studies are needed that involve large data sets, various pathologies, and other MR sequences of the lumbar spine.
Notes
AUTHOR CONTRIBUTIONS
Conceptualization : JHL, SJ, CBJ, IHH
Data curation : ISL, YSS, DHK, SY
Formal analysis : JHL, JIL
Funding acquisition : IHH, JIL
Methodology : CBJ, HK
Project administration : SJ, HK
Visualization : SJ, CBJ
Writing - original draft : CBJ, JHL
Writing - review & editing : CBJ, IHH
References
1. Beechar VB, Zinn PO, Heck KA, Fuller GN, Han I, Patel AJ, et al. Spinal epidermoid tumors: case report and review of the literature. Neurospine. 15:117–122. 2018.

2. Bi L, Kim J, Kumar A, Feng D, Fulham M. Synthesis of Positron Emission Tomography (PET) Images via Multi-channel Generative Adversarial Networks (GANs). In : Cardoso MJ, Arbel T, editors. Molecular Imaging, Reconstruction and Analysis of Moving Body Organs, and Stroke Imaging and Treatment. Cham: Springer;2017. p. 43–51.
3. Creswell A, White T, Dumoulin V, Arulkumaran K, Sengupta B, Bharath AA. Generative adversarial networks: an overview. IEEE Signal Processing Magazine. 35:53–65. 2018.

4. Feng R, Badgeley M, Mocco J, Oermann EK. Deep learning guided stroke management: a review of clinical applications. J Neurointerv Surg. 10:358–362. 2018.

5. Goodfellow IJ, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, et al. Generative adversarial nets. Adv Neural Inf Process Syst. 27:2672–2680. 2014.
6. He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In : Proc IEEE Int Conf Comput Vis; p. 770–778. 2016.

7. Hirasawa T, Aoyama K, Tanimoto T, Ishihara S, Shichijo S, Ozawa T, et al. Application of artificial intelligence using a convolutional neural network for detecting gastric cancer in endoscopic images. Gastric Cancer. 21:653–660. 2018.

8. Isola P, Zhu JY, Zhou T, Efros AA. Image-to-image translation with conditional adversarial networks. In : Proc IEEE Int Conf Comput Vis; p. 1125–1134. 2017.

9. Jin CB, Kim H, Jung W, Joo S, Park E, Saem AY, et al. Deep CT to MR synthesis using paired and unpaired data. Sensors (Basel). 19:2019. 2019.

10. Johnson J, Alahi A, Fei-Fei L. Perceptual Losses for Real-Time Style Transfer and Super-Resolution. In : Leibe B, Matas J, Sebe N, Welling M, editors. Computer Vision - ECCV 2016. Cham: Springer;2016. p. 694–711.
11. Kingma DP, Ba J. Adam: a method for stochastic optimization. Available at : https://arxiv.org/abs/1412.6980.
12. Nie D, Trullo R, Lian J, Petitjean C, Ruan S, Wang Q, et al. Medical image synthesis with context-aware generative adversarial networks. Med Image Comput Comput Assist Interv. 10435:417–425. 2017.

13. Olczak J, Fahlberg N, Maki A, Razavian AS, Jilert A, Stark A, et al. Artificial intelligence for analyzing orthopedic trauma radiographs. Acta Orthop. 88:581–586. 2017.

14. Pathak D, Krahenbuhl P, Donahue J, Darrell T, Efros AA. Context encoders: feature learning by inpainting. In : Proc IEEE Int Conf Comput Vis; p. 2536–2544. 2016.

15. Ulyanov D, Vedaldi A, Lempitsky V. Instance normalization: the missing ingredient for fast stylization. Available at : https://arxiv.org/abs/1607.08022.
16. Wolterink JM, Dinkla AM, Savenije MH, Seevinck PR, van den Berg CA, Išgum I. Deep MR to CT synthesis using unpaired data. Available at : https://arxiv.org/abs/1708.01155.

17. Xu B, Wang N, Chen T, Li M. Empirical evaluation of rectified activations in convolutional network. Available at : https://arxiv.org/abs/1505.00853.
Fig. 1.
Flow diagram of the deep generative adversarial networks (GANs). CT : computed tomography, MR : magnetic resonance.
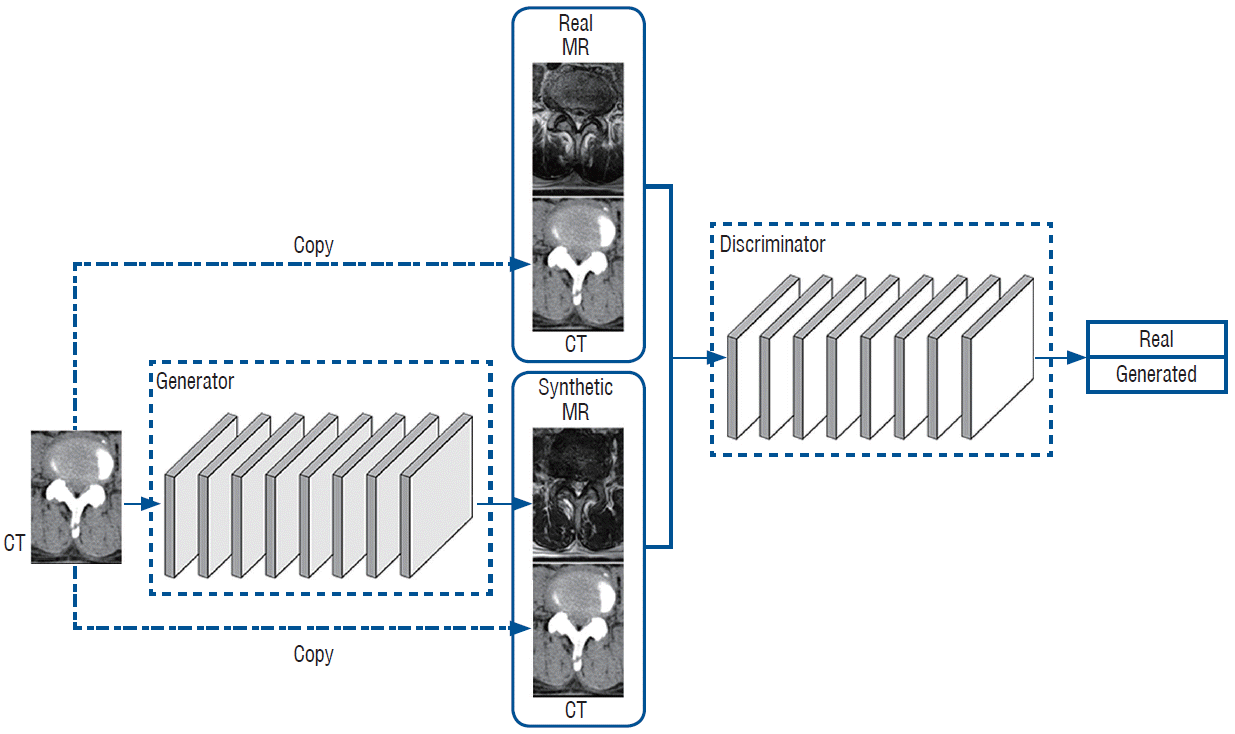
Fig. 2.
Selecting the MR axial image corresponding to the CT image. Top : the axial planes of the sagittal MR and CT were parallel. Axial images at this disc level were included. Bottom : the axial plane of the sagittal MR and CT images were not parallel. This paired images were excluded. CT : computed tomography, MR : magnetic resonance.
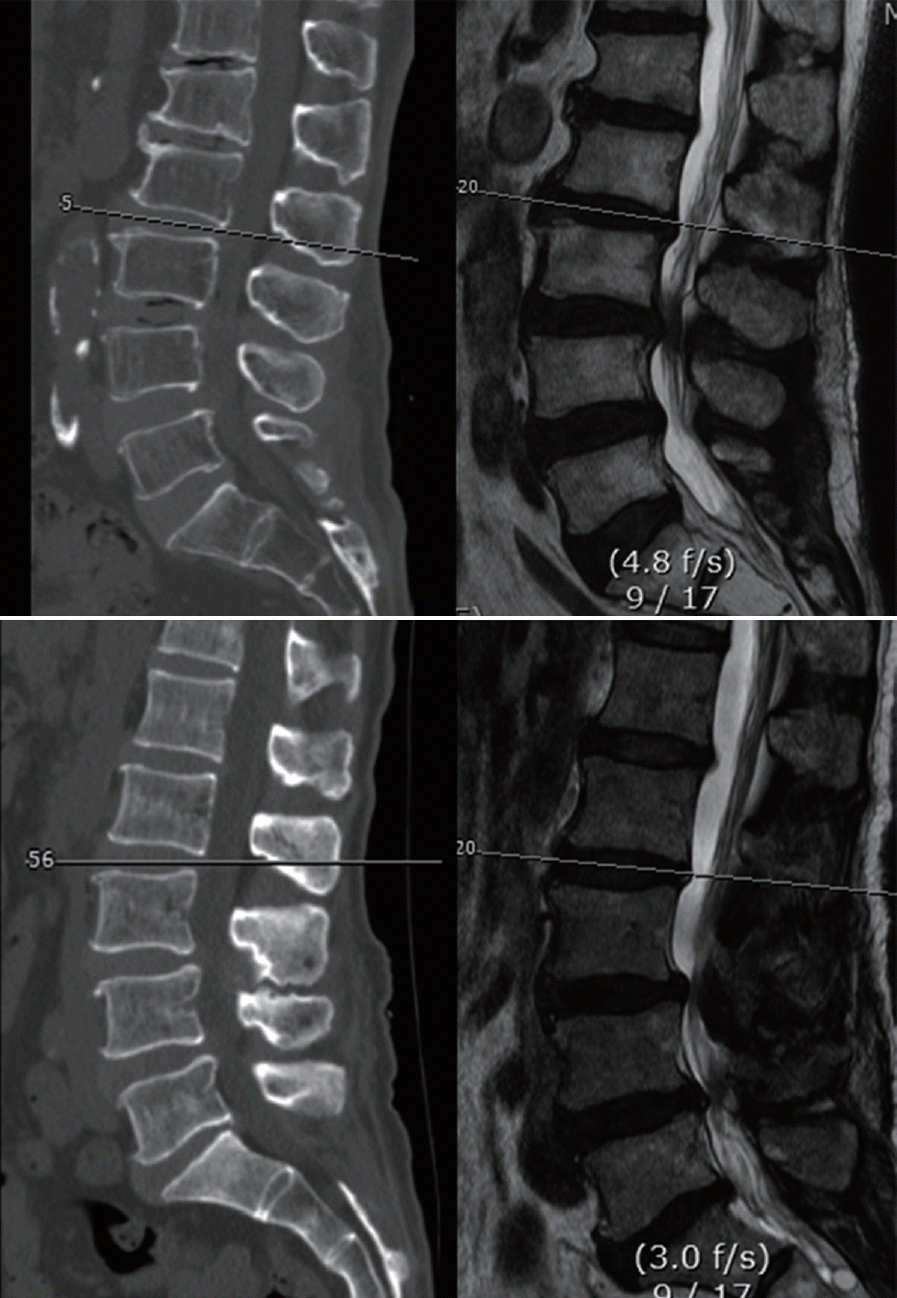
Fig. 3.
Range of image cropping. Horizontally, the image was cut parallel at the most ventral part of the lumbar vertebral body. It was then cut in the dorsal direction from the center of thecal sac to the ventral end of the vertebral body (left side). Finally, it was cut vertically by the same length at the center of thecal sac (right side). The range within which both lateral sides of the vertebral body could be seen in all images was measured. This range was then applied to all images.
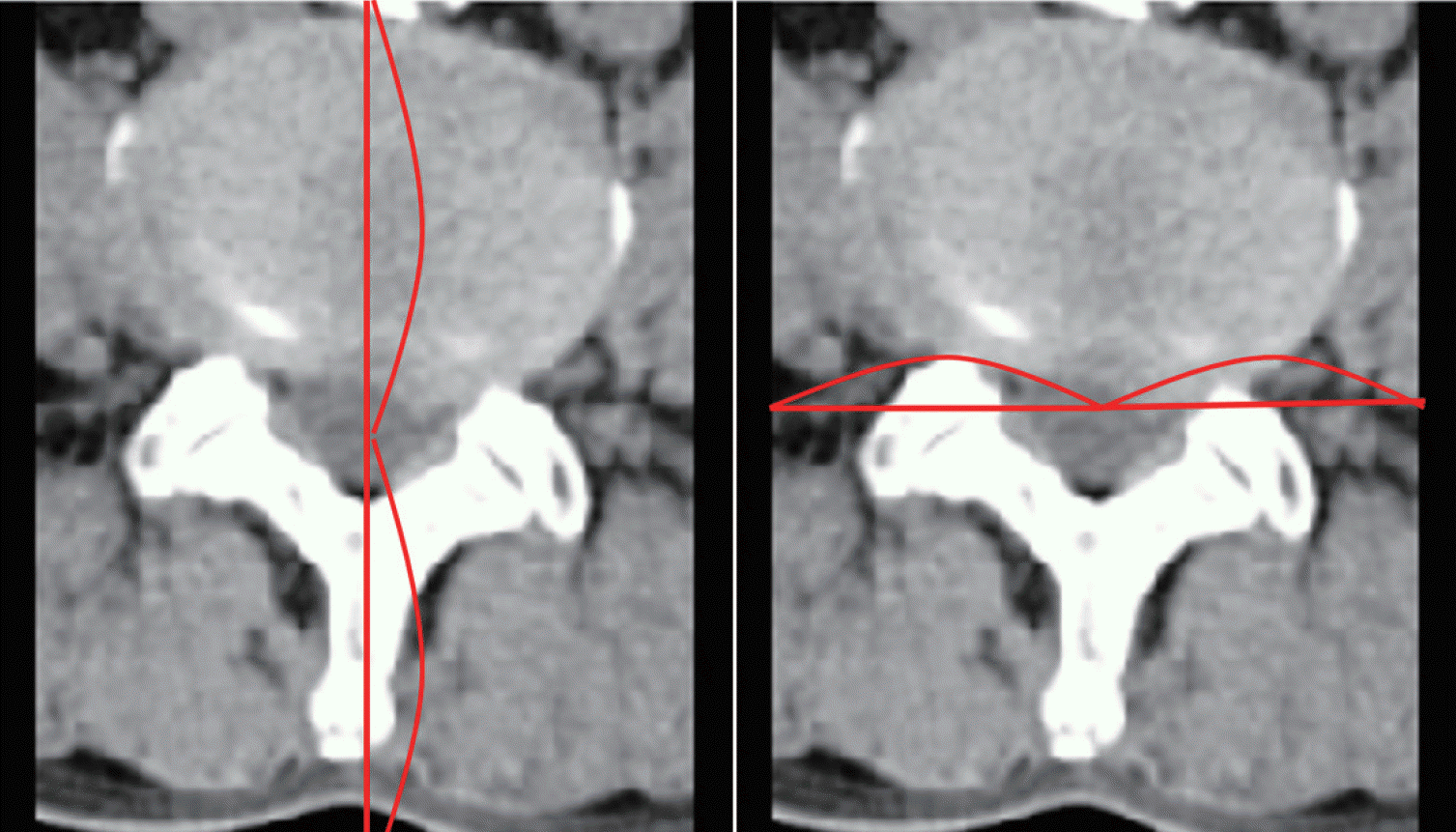
Fig. 4.
Qualitative comparison between synthetic MR and real MR images. Questionnaires were given to six medical doctors (two radiologists, two spine surgeons, and two neurosurgical residents) who had not seen synthetic MR images before. They were asked to select the real MR image. CT : computed tomography, MR : magnetic resonance.

Fig. 5.
Evaluation of the similarities between synthetic MR and real MR images by radiologists. The similarity of each structure showed in spine CT and MR images was evaluated in terms of percentile by two radiologists. CT : computed tomography, MR : magnetic resonance.

Fig. 6.
All CT scans (CT01–CT15) to generate the synthetic magnetic resonance T2 weighted axial (MRT2) images were listed. The left side shows the CT scans, the middle displays the synthetic MRT2 images, and the right side comprises the real MRT2 images. CT : computed tomography.

Fig. 7.
Examples of MAE between the real and synthesized MR images. MAE : mean absolute error, SynMR : synthetic magnetic resonance image, IMR : real magnetic resonance image, MR : magnetic resonance.

Table 1.
Quantitative evaluation with MAE and PSNR
Table 2.
Results of selection between real and synthetic MR images corresponding to spinal CT images
Table 3.
Similarity between real and synthetic MR images analyzed by expert radiologists (similarities are listed in rad01–rad02 order)




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download