INTRODUCTION
Carbapenem-resistant Enterobacteriaceae (CRE) has emerged as a major resistance concern over the past decade (1), with reports of extremely high prevalence worldwide (2, 3, 4) and a high potential for rapid transmission (5). The global emergence of carbapenemase-producing Enterobacteriaceae (CPE) poses a threat to the achievements of modern medicine (6) and represents an increasing threat to patient safety in the world (7). Carbapenemases are a group of β-lactamases that are capable of hydrolyzing carbapenem antibiotics, in addition to cephalosporins and other β-lactam antimicrobials (8). Three major classes of carbapenemases is widespread globally in clinical CRE clinical isolates, such as class A (mainly KPC[Klebsiella pneumoniae carbapenemases]), class B (VIM[Verona integron-encoded metallo-β-lactamase], NDM[New Delhi metallo-β-lactamase], and IMP[Imipenemase metallo-β-lactamase]) and class D (OXA[Oxacillinase]-48 and its variants, OXA-162, and OXA-181, etc.) (8, 9, 10). The carbapenemase genes in Enterobacteriaceae have been shown to be associated with clonal spread and plasmid-mediated transmission contribute to the ongoing rise in the incidence of these bacteria (11). CRE outbreak in hospitals has become a critical issue and transmission by patients with CRE, as well as carriers of CRE, contribute significantly to in-hospital CRE transmission (12). In Korea, CRE was first reported in 2010 and has been continuously increasing afterward. However, domestic clinical reports on related CRE and CPE are still very limited (13, 14). Therefore, this study investigated the prevalence and characteristics of CPE among CRE strains isolated from Korean adult and pediatric patients.
MATERIALS AND METHODS
Bacteria strains
A total of 8,147 clinical isolates were obtained from blood, urine, stool, sputum, lesion, bile, pus, tracheal aspiration, and etc., hospitals from 2018 to 2020.
Microbial characterization
Bacteria characterization was performed as described previously (15). Species were identified by Bruker Biotyper MALDI-TOF MS (Bruker Daltonics GmbH; Bremen, Germany) and VITEK 2 (BioMérieux; Marcy l'Etoile, France). Antibiotic resistance test were performed with TREK Diagnostic System (Cleveland, OH, USA). According to Clinical and Laboratory Standards Institute (CLSI) guidelines, CRE was resistant to imipenem, meropenem, doripenem, or ertapenem (16).
Polymerase chain reaction (PCR) for identifying CPE genes
Major CPE genes, blaKPC and blaGES from class A; blaIMP, blaVIM, and blaNDM from class B; and blaOXA from class D, was detected by using PCR as described previously (17). Primer used in this study was listed in Table 1.
Table 1.
Primer pairs used for amplification of CPE
RESULTS
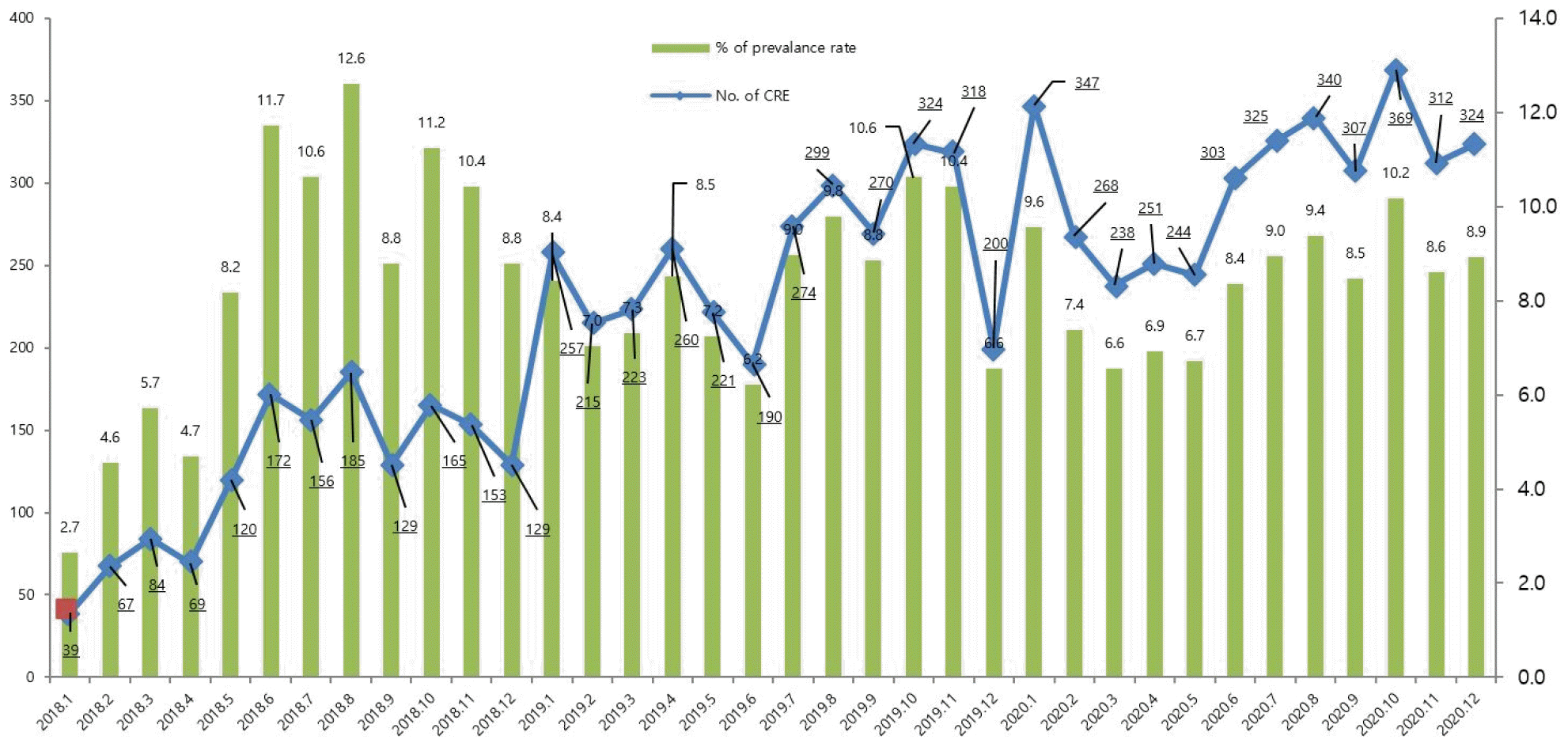
We first analyzed the occurrence pattern of CREs monthly between 2018 to 2020 (Fig. 1) For two years, the occurrence of CRE showed a monthly increase pattern, and as a representative example, it was confirmed that it increased more than 10 times in January 2020 (n=347; 9.6% of prevalence rate) compared to January 2018 (n=39; 2.7% of prevalence rate). The highest month was October 2020 (n=369) and the lowest month was January 2018 (n=39).
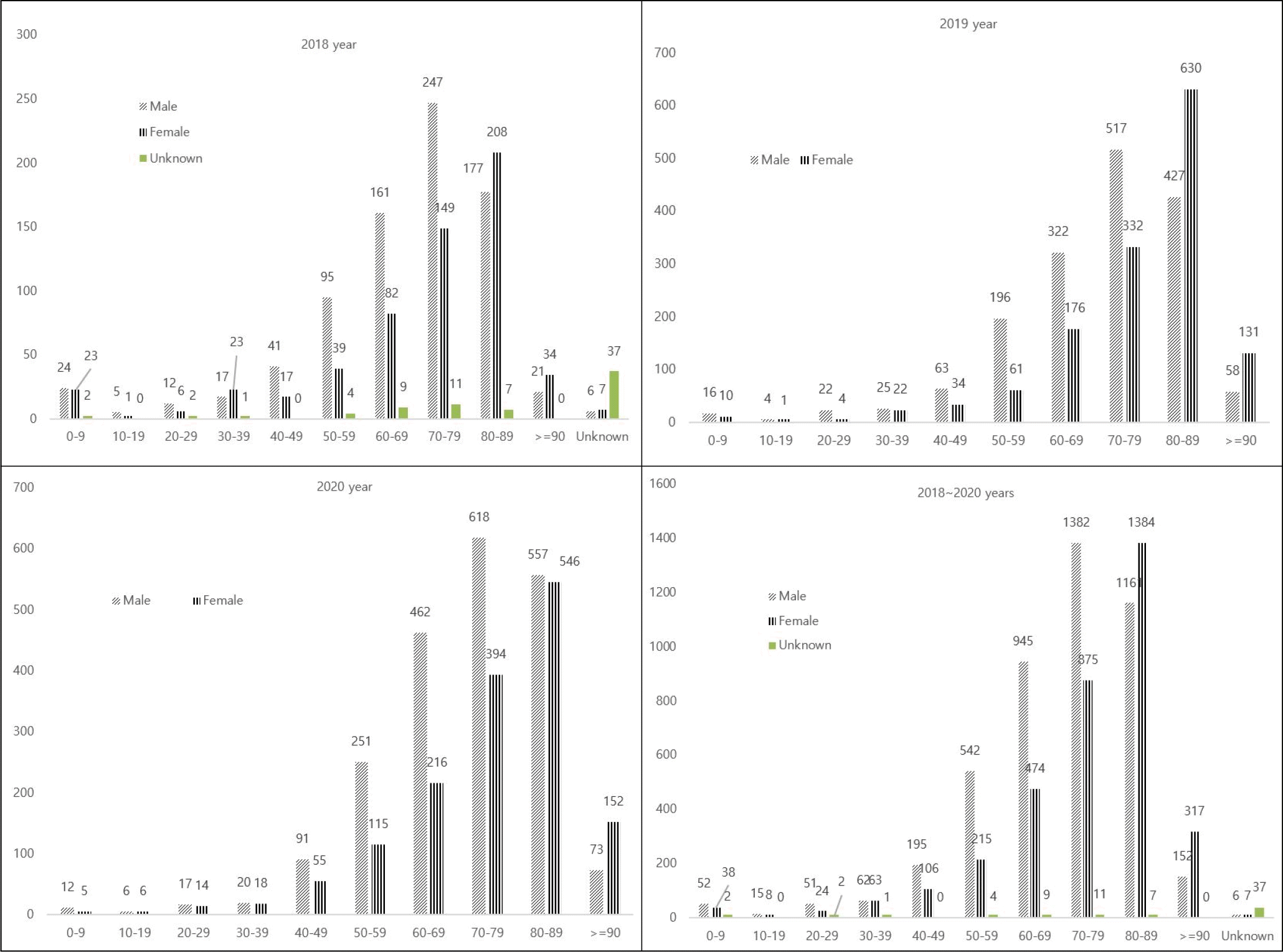
Next, we analyzed age and gender distribution yearly. As shown in Fig. 2, most infections occurred in adults over 60 years of age (n=6,767, 83.1%). In addition, the infection rate was higher in men under 80 years of age than in women, but it was similar in those over 80 years of age.
Fig. 2
Distribution of CRE by gender and age group between 2018 and 2020 in Seoul, Korea. X axis (Age), Y axis (Numbers).
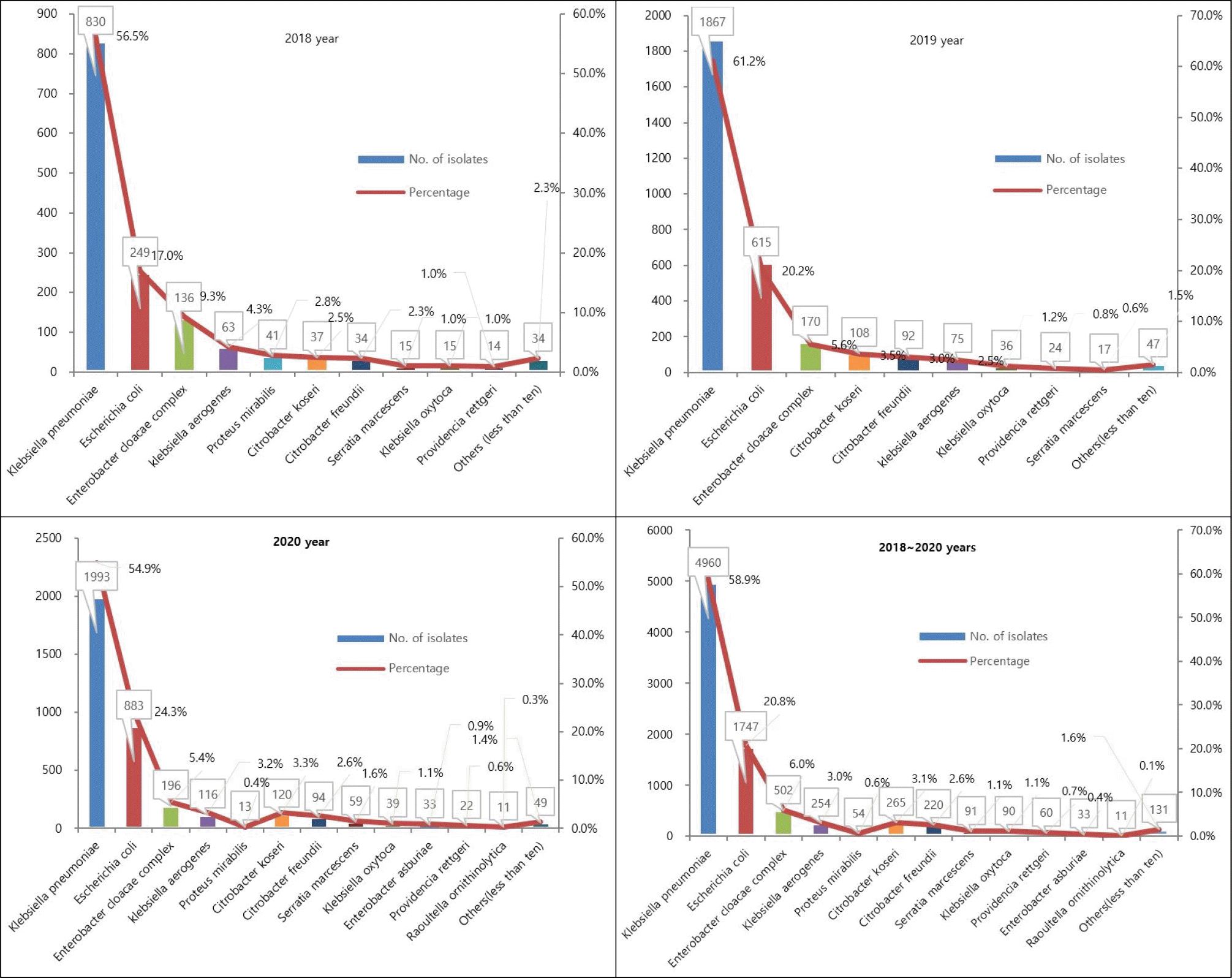
As a result of analyzing the species of all CRE clinical isolates (n=8,147), K. pneumoniae was the most common with 4,690 strains (58.9%). E. coli was the second most common species with 1,747 isolates (20.8%). These two species were found equally the most in all years (Fig. 3).
Fig. 3
Distribution of CRE by species between 2018 and 2020 in Seoul, Korea. X axis (Species), Y left axis (Numbers).
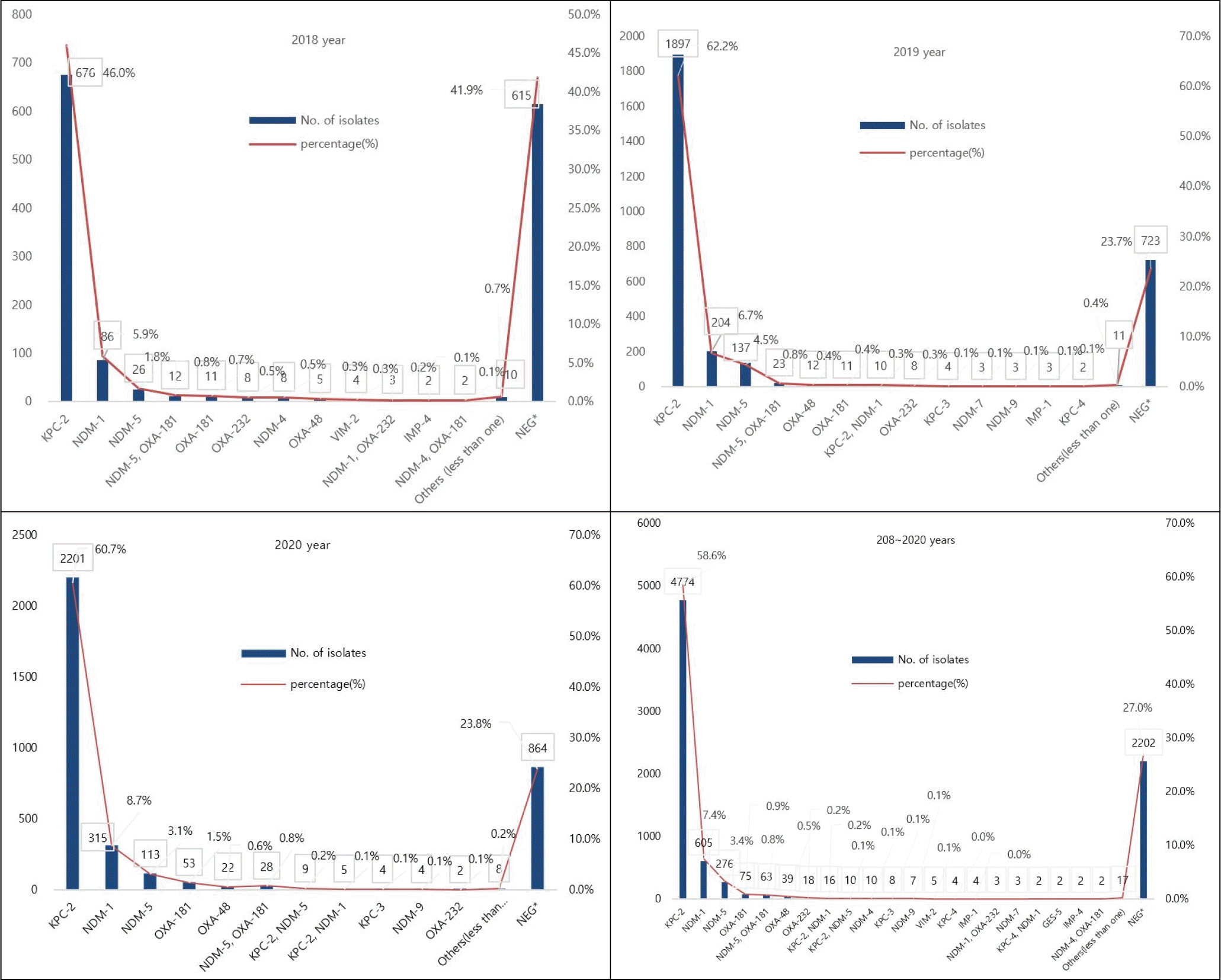
The types of carbapenemases observed in CRE isolates were analyzed using PCR (Fig. 4). Positive for one or more CPE types was detected in 5,945 isolates (63.0%), and among the CPE types, KPC-2 (58.6%), NDM-1 (7.4%), NDM-5 (3.4%) were detected. Minor CPE types was NDM-5 (1%), OXA-181(0.7%), OXA-181(0.9%), OXA-48(0.5%), OXA-232 (0.2%), KPC-2 with NDM-1 (0.2%), and KPC-2 with NDM-5 (0.1%), NDM-4 (0.1%) VIM-2 (0.1%), and KPC-3 (0.1%). Coexistence of both NDM-5 and OXA-181 was detected in E. coli (60/63, 95.2%) and K. pneumoniae (3/63, 4.8%).
Fig. 4
Distribution of CPE genotypes between 2018 and 2020 in Seoul, Korea. X axis (Types), Y left axis (Numbers).
KPC-2 (3,428/8,147isolates [42.1%]) and NDM-1 (156/8,147isolates [1.9%]), which represented the majority of CPE are most commonly found in K. pneumoniae, while the next most frequently found NDM-5 (244/8,147isolates [3.0%]) and OXA-181 (57/8,147isolates [0.7%]) are mostly found in E. coli (Table 2). Co-existence of NDM-5 and OXA-181 was detected in E. coli (60/63, 95.2%) followed by K. pneumoniae (3/63, 4.8%).
Table 2.
Distributions of genotypes of carbapenemase-producing Enterobacteriaceae isolates during 3 years (between 2018 and 2020)
DISCUSSION
CRE is an important public health concern for both worldwide and South Korea, which is a major problem for patient treatment and infection control (18). Since it was first reported in 2010, the incidence of CRE has been increasing in Korea (19). This study describes the prevalence and characteristics of CP-CREs (carbapenemase producing carbapenem nonsusceptible Enterobacteriaceaes) in Korea.
There are many factors that contribute to the increased incidence of CRE (Fig. 1) Recent studies have found that the increasing trend of CRE infection was estimated to be due to various factors, such as an increase in reporting institutions after the transition to a mandatory surveillance system, an increase in the actual occurrence of CRE infection, and an increase in screening tests for high-risk groups at medical institutions (20). A similar pattern was observed in report of total surveillance reported through the Korea Disease Control and Prevention Agency's (KDCA) surveillance system from 2018 to 2020 (20).
The ages of patients infected with CREs ranged from under 1 year of age to 90 years. Most patients were older than 60 years (6,672 of 8,147, 81.8%) but this reflects the bias in reporting and sampling, since most of the cases reported during this period were adults over the age of 60s.
In our study, male gender was a risk factor for CPE carriage. This agrees with previous studies that demonstrated associations between male gender and methicillin-resistant Staphylococcus aureus (MRSA), as well as health care-associated infections (21). Previous studies have shown that male gender is also a risk factor for CPE, specifically, NDM producing Enterobacteriaceae (22). The reason for such a low prevalence in women could be explained by gender differences in hygiene notions, suggesting that women show better compliance to hand hygiene and soap use than men in Community studies (23, 24).
Our findings that E. coli and K. pneumoniae represented the most frequently isolated CRE agree with those of previous studies of epidemiology of carbapenemase-producing Enterobacteriaceae in hospitalized patients in Italy (25), USA (26, 27, 28), and are most often mediated by CPE which can be shared across species (29). Clinically significant carbapenem resistance occurs across Enterobacterales spp., particularly Klebsiella spp., E. coli and Enterobacter spp. (26, 27, 30), and is most often mediated by carbapenemase genes which can be shared across species (29).
CPE represent an increasing global threat worldwide and K. pneumoniae carbapenemase-producing K. pneumoniae (KPC-KP) has become one of the most important contemporary pathogens, especially in endemic areas (31). K. pneumoniae carbapenemase (KPC, encoded by blaKPC) is one of the most common carbapenemase genes globally (32). KPC was previously described as the most common type of carbapenemase in K. pneumoniae strains in adults (33), which is consistent with the current results that the 59.2% (4,821 of 8,147) KPCs collected from patients mainly produced the KPC-2 (99%; 4,774 of 4,821) of KPC enzyme.
KPC is the most common carbapenemase worldwide, but NDM has been shown to be a significant sporadic outbreak worldwide (34, 35). Since initial isolation from clinical isolates of K. pneumoniae in 2008 (36), NDM-1 is of major concern because of many human and biological factors leading to a wide dissemination of blaNDM-1-carrying strains, especially K. pneumoniae (37, 38). Furthermore, travel directly from India and Iran since the outbreak in Greece in 2014 is also related to the first case of NDM isolation in the United States (39).
The analysis of a total of 8,147 isolates of Enterobacteriaceae for the presence of CPE demonstrated NDM as the second gene (11.1%) and NDM-1 (67.1%, 605/902) was found to be the most predominant NDM gene of the Enterobacteriaceae in this study. The accumulated data from previous studies and this study indicate that NDM-1 enzymes gradually became the most prevalent type of carbapenemase in China and various European regions, including the Mediterranean basin and the Balkan region of Carbapenem-resistant K. pneumoniae (CRKP) strains (7, 40, 41, 42, 43, 44). NDM-positive strains are continuing to spread worldwide despite continuous efforts and remain a critical challenge for clinical treatment and a significant threat to public health (45).
In Korea, KPC-producing strains were first reported in K. pneumoniae in 2010 (46), and NDM-1-producing strains were first isolated from K. pneumoniae in 2010 and reported in 2012 (47). Since then, the incidence has continued to increase, and CPE outbreaks are intermittently reported (48). To date, there have been several reports of Korea are famous for the outbreak of K. pneumoniae, which produces KPC-2, NDM-1, and OXA-232 (47, 49, 50). Most of the types of CPE detected in this study were KPC-2 (58.6%), followed by NDM-1 (7.4%), and NDM-5 (3.4%), suggesting that KPC and NDM are widespread in Seoul, Korea.
The co-existence detection rate of NDM-5 and OXA-181 was 0.7% (63/8,147), showing a similar level every year. In particular, blaNDM and blaOXA, which co-produce the Enterobacteriaceae, are emerging as serious challenges to treatment, infection control, and public health (51, 52, 53). Our findings that co-producing of NDM-5 and OXA-181 was detected in E. coli followed by K. pneumoniae are in agreement with those of previous studies of various clinical specimens in Myanmar and India (52, 53). Similarly, in Korea, a blaNDM-5 and blaOXA-181 Co-producing K. pneumoniae strain was first detected in 2014 (54), and by 2017, which is hospital witnessed the first case caused by blaNDM-5 and blaOXA-48 co-producing uropathogenic E. coli (55). As shown in this study, the emergence of extremely drug-resistant isolates with multiple carbapenemase genes requires collaboration on appropriate infection control measures for this, due to limited treatment options and potential global spread through cross-border transfer (54).
These findings provide good basic data for comprehensive surveillance of CREs in Seoul, and the molecular characterization of CREs isolated from patients and the epidemiology of infection is essential for developing of effective prevention strategies.




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download